[English] 日本語
 Yorodumi
Yorodumi- EMDB-7381: Single Molecule 3D Image of a Hole-Hole Homodimer Species Generat... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7381 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single Molecule 3D Image of a Hole-Hole Homodimer Species Generated in the Production of a Bispecific Recombinant IgG1 (No. 029) | |||||||||||||||
 Map data Map data | Single Molecule 3D Image of a Hole-Hole Homodimer Species Generated in the Production of a Bispecific Recombinant IgG1 (No. 029) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species | unidentified (others) | |||||||||||||||
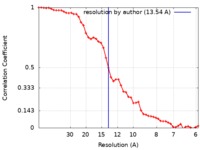
| Method | electron tomography / negative staining / Resolution: 13.54 Å | |||||||||||||||
 Authors Authors | Lei D / Liu J / Liu H / Lei M / Ren G | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2019 Journal: Sci Rep / Year: 2019Title: Single-Molecule 3D Images of "Hole-Hole" IgG1 Homodimers by Individual-Particle Electron Tomography. Authors: Dongsheng Lei / Jianfang Liu / Hongbin Liu / Thomas E Cleveland / John P Marino / Ming Lei / Gang Ren /  Abstract: The engineering of immunoglobulin-G molecules (IgGs) is of wide interest for improving therapeutics, for example by modulating the activity or multiplexing the specificity of IgGs to recognize more ...The engineering of immunoglobulin-G molecules (IgGs) is of wide interest for improving therapeutics, for example by modulating the activity or multiplexing the specificity of IgGs to recognize more than one antigen. Optimization of engineered IgG requires knowledge of three-dimensional (3D) structure of synthetic IgG. However, due to flexible nature of the molecules, their structural characterization is challenging. Here, we use our reported individual-particle electron tomography (IPET) method with optimized negative-staining (OpNS) for direct 3D reconstruction of individual IgG hole-hole homodimer molecules. The hole-hole homodimer is an undesired variant generated during the production of a bispecific antibody using the knob-into-hole heterodimer technology. A total of 64 IPET 3D density maps at ~15 Å resolutions were reconstructed from 64 individual molecules, revealing 64 unique conformations. In addition to the known Y-shaped conformation, we also observed an unusual X-shaped conformation. The 3D structure of the X-shaped conformation contributes to our understanding of the structural details of the interaction between two heavy chains in the Fc domain. The IPET approach, as an orthogonal technique to characterize the 3D structure of therapeutic antibodies, provides insight into the 3D structural variety and dynamics of heterogeneous IgG molecules. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7381.map.gz emd_7381.map.gz | 6.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7381-v30.xml emd-7381-v30.xml emd-7381.xml emd-7381.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7381_fsc.xml emd_7381_fsc.xml | 4.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_7381.png emd_7381.png | 13.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7381 http://ftp.pdbj.org/pub/emdb/structures/EMD-7381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7381 | HTTPS FTP |
-Related structure data
| Related structure data |  7353C  7354C  7355C  7356C  7357C  7358C  7359C  7360C  7361C  7362C  7363C  7364C  7365C  7366C  7367C  7368C  7369C  7370C  7371C  7372C  7373C  7374C  7375C  7376C  7377C  7378C  7379C  7380C  7382C  7383C  7384C  7385C  7386C  7387C  7388C  7389C  7390C  7391C  7392C  7393C  7394C  7395C  7396C  7397C  7398C  7399C  7400C  7401C  7402C  7404C  7405C  7406C  7407C  7408C  7409C  7410C  7411C  7412C 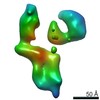 7413C  7414C  7415C  7416C  7417C  7418C  7419C  7420C  7421C  7422C  7423C 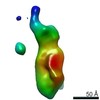 7424C  7425C  7426C 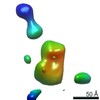 7427C  7428C  7429C  7430C  7431C  7432C  7433C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7381.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7381.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single Molecule 3D Image of a Hole-Hole Homodimer Species Generated in the Production of a Bispecific Recombinant IgG1 (No. 029) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.96 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : IgG1 hole-hole homodimer
| Entire | Name: IgG1 hole-hole homodimer |
|---|---|
| Components |
|
-Supramolecule #1: IgG1 hole-hole homodimer
| Supramolecule | Name: IgG1 hole-hole homodimer / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism: unidentified (others) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: Dulbecco's phosphate buffered saline (DPBS) |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate / Details: The grid was stained with 1% (w/v) uranyl formate |
| Grid | Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa |
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | ZEISS LIBRA120PLUS |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 41.74 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal magnification: 80000 |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)