[English] 日本語
 Yorodumi
Yorodumi- EMDB-7336: The Cryo-EM structure of a eukaryotic oligosaccharyl transferase ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7336 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
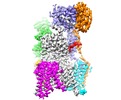
| Title | The Cryo-EM structure of a eukaryotic oligosaccharyl transferase complex | |||||||||
 Map data Map data | Eukaryotic oligosaccharyl transferase complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / TRANSFERASE / glycosylation | |||||||||
| Function / homology |  Function and homology information Function and homology informationMiscellaneous transport and binding events / dolichol-linked oligosaccharide biosynthetic process / protein O-linked glycosylation via mannose / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / oligosaccharyltransferase complex / : / : / protein N-linked glycosylation / glycosyltransferase activity ...Miscellaneous transport and binding events / dolichol-linked oligosaccharide biosynthetic process / protein O-linked glycosylation via mannose / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / oligosaccharyltransferase complex / : / : / protein N-linked glycosylation / glycosyltransferase activity / protein-disulfide reductase activity / Neutrophil degranulation / post-translational protein modification / nuclear envelope / protein-macromolecule adaptor activity / endoplasmic reticulum membrane / structural molecule activity / endoplasmic reticulum / mitochondrion / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Bai L / Li H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: The atomic structure of a eukaryotic oligosaccharyltransferase complex. Authors: Lin Bai / Tong Wang / Gongpu Zhao / Amanda Kovach / Huilin Li /  Abstract: N-glycosylation is a ubiquitous modification of eukaryotic secretory and membrane-bound proteins; about 90% of glycoproteins are N-glycosylated. The reaction is catalysed by an eight-protein ...N-glycosylation is a ubiquitous modification of eukaryotic secretory and membrane-bound proteins; about 90% of glycoproteins are N-glycosylated. The reaction is catalysed by an eight-protein oligosaccharyltransferase (OST) complex that is embedded in the endoplasmic reticulum membrane. Our understanding of eukaryotic protein N-glycosylation has been limited owing to the lack of high-resolution structures. Here we report a 3.5 Å resolution cryo-electron microscopy structure of the Saccharomyces cerevisiae OST complex, revealing the structures of subunits Ost1-Ost5, Stt3, Wbp1 and Swp1. We found that seven phospholipids mediate many of the inter-subunit interactions, and an Stt3 N-glycan mediates interactions with Wbp1 and Swp1 in the lumen. Ost3 was found to mediate the OST-Sec61 translocon interface, funnelling the acceptor peptide towards the OST catalytic site as the nascent peptide emerges from the translocon. The structure provides insights into co-translational protein N-glycosylation, and may facilitate the development of small-molecule inhibitors that target this process. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7336.map.gz emd_7336.map.gz | 8.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7336-v30.xml emd-7336-v30.xml emd-7336.xml emd-7336.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7336.png emd_7336.png | 163.7 KB | ||
| Filedesc metadata |  emd-7336.cif.gz emd-7336.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7336 http://ftp.pdbj.org/pub/emdb/structures/EMD-7336 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7336 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7336 | HTTPS FTP |
-Related structure data
| Related structure data |  6c26MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7336.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7336.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Eukaryotic oligosaccharyl transferase complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.088 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
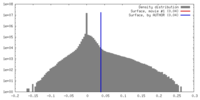
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : oligosaccharyl transferase complex
+Supramolecule #1: oligosaccharyl transferase complex
+Macromolecule #1: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #2: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #3: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #4: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #5: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #6: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #7: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #8: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #11: (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)met...
+Macromolecule #12: 2-acetamido-2-deoxy-beta-D-glucopyranose
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6c26: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)