+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-7325 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation | |||||||||
 マップデータ マップデータ | b-factor sharpening 100 | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | ABC transporter / ABCB1 / P-glycoprotein / cryo-EM / multidrug resistance / Structural Genomics / PSI-2 / Protein Structure Initiative / TRANSPORT PROTEIN | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報carboxylic acid transmembrane transport / carboxylic acid transmembrane transporter activity / hormone transport / cellular response to nonylphenol / cellular response to borneol / response to codeine / response to cyclosporin A / cellular response to mycotoxin / daunorubicin transport / positive regulation of response to drug ...carboxylic acid transmembrane transport / carboxylic acid transmembrane transporter activity / hormone transport / cellular response to nonylphenol / cellular response to borneol / response to codeine / response to cyclosporin A / cellular response to mycotoxin / daunorubicin transport / positive regulation of response to drug / terpenoid transport / ceramide floppase activity / negative regulation of sensory perception of pain / positive regulation of establishment of Sertoli cell barrier / regulation of intestinal absorption / cellular response to external biotic stimulus / response to quercetin / response to antineoplastic agent / ceramide translocation / floppase activity / Abacavir transmembrane transport / establishment of blood-retinal barrier / phosphatidylethanolamine flippase activity / protein localization to bicellular tight junction / phosphatidylcholine floppase activity / external side of apical plasma membrane / Atorvastatin ADME / xenobiotic transport across blood-brain barrier / response to thyroxine / establishment of blood-brain barrier / transepithelial transport / xenobiotic detoxification by transmembrane export across the plasma membrane / export across plasma membrane / P-type phospholipid transporter / cellular response to L-glutamate / ABC-type xenobiotic transporter / response to vitamin A / response to vitamin D / response to glycoside / response to alcohol / response to glucagon / intestinal absorption / ABC-type xenobiotic transporter activity / Prednisone ADME / cellular response to antibiotic / phospholipid translocation / cellular hyperosmotic salinity response / maintenance of blood-brain barrier / cellular response to alkaloid / efflux transmembrane transporter activity / transmembrane transporter activity / xenobiotic transmembrane transporter activity / ATPase-coupled transmembrane transporter activity / cellular response to dexamethasone stimulus / response to cadmium ion / transport across blood-brain barrier / lactation / xenobiotic metabolic process / regulation of chloride transport / response to progesterone / placenta development / stem cell proliferation / cellular response to estradiol stimulus / brush border membrane / female pregnancy / circadian rhythm / ABC-family proteins mediated transport / transmembrane transport / G2/M transition of mitotic cell cycle / cellular response to tumor necrosis factor / cellular response to lipopolysaccharide / response to hypoxia / apical plasma membrane / response to xenobiotic stimulus / ubiquitin protein ligase binding / cell surface / ATP hydrolysis activity / extracellular exosome / ATP binding / membrane / plasma membrane / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.4 Å | |||||||||
 データ登録者 データ登録者 | Kim YJ / Chen J | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Science / 年: 2018 ジャーナル: Science / 年: 2018タイトル: Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. 著者: Youngjin Kim / Jue Chen /  要旨: The multidrug transporter permeability (P)-glycoprotein is an adenosine triphosphate (ATP)-binding cassette exporter responsible for clinical resistance to chemotherapy. P-glycoprotein extrudes toxic ...The multidrug transporter permeability (P)-glycoprotein is an adenosine triphosphate (ATP)-binding cassette exporter responsible for clinical resistance to chemotherapy. P-glycoprotein extrudes toxic molecules and drugs from cells through ATP-powered conformational changes. Despite decades of effort, only the structures of the inward-facing conformation of P-glycoprotein are available. Here we present the structure of human P-glycoprotein in the outward-facing conformation, determined by cryo-electron microscopy at 3.4-angstrom resolution. The two nucleotide-binding domains form a closed dimer occluding two ATP molecules. The drug-binding cavity observed in the inward-facing structures is reorientated toward the extracellular space and compressed to preclude substrate binding. This observation indicates that ATP binding, not hydrolysis, promotes substrate release. The structure evokes a model in which the dynamic nature of P-glycoprotein enables translocation of a large variety of substrates. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_7325.map.gz emd_7325.map.gz | 200.1 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-7325-v30.xml emd-7325-v30.xml emd-7325.xml emd-7325.xml | 18.2 KB 18.2 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_7325.png emd_7325.png | 46.2 KB | ||
| Filedesc metadata |  emd-7325.cif.gz emd-7325.cif.gz | 6.7 KB | ||
| その他 |  emd_7325_additional.map.gz emd_7325_additional.map.gz | 15.8 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7325 http://ftp.pdbj.org/pub/emdb/structures/EMD-7325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7325 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_7325_validation.pdf.gz emd_7325_validation.pdf.gz | 440.1 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_7325_full_validation.pdf.gz emd_7325_full_validation.pdf.gz | 439.6 KB | 表示 | |
| XML形式データ |  emd_7325_validation.xml.gz emd_7325_validation.xml.gz | 7.1 KB | 表示 | |
| CIF形式データ |  emd_7325_validation.cif.gz emd_7325_validation.cif.gz | 8.1 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7325 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7325 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7325 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7325 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  6c0vMC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | |
| 電子顕微鏡画像生データ |  EMPIAR-10803 (タイトル: Cryo-electron microscopy reconstruction of ATP-bound human P-glycoprotein EMPIAR-10803 (タイトル: Cryo-electron microscopy reconstruction of ATP-bound human P-glycoproteinData size: 2.0 TB Data #1: Unaligned and uncorrected multiframe movies of human ATP-bound P-glycoprotein [micrographs - multiframe]) |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_7325.map.gz / 形式: CCP4 / 大きさ: 216 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_7325.map.gz / 形式: CCP4 / 大きさ: 216 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | b-factor sharpening 100 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-追加マップ: original map
| ファイル | emd_7325_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | original map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
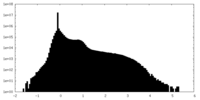
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Human P-glycoprotein E556Q, E1201Q
| 全体 | 名称: Human P-glycoprotein E556Q, E1201Q |
|---|---|
| 要素 |
|
-超分子 #1: Human P-glycoprotein E556Q, E1201Q
| 超分子 | 名称: Human P-glycoprotein E556Q, E1201Q / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 141 KDa |
-分子 #1: Multidrug resistance protein 1
| 分子 | 名称: Multidrug resistance protein 1 / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO / EC番号: ec: 3.6.3.44 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 142.660922 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: MDLEGDRNGG AKKKNFFKLN NKSEKDKKEK KPTVSVFSMF RYSNWLDKLY MVVGTLAAII HGAGLPLMML VFGEMTDIFA NAGNLEDLM SNITNRSDIN DTGFFMNLEE DMTRYAYYYS GIGAGVLVAA YIQVSFWCLA AGRQIHKIRK QFFHAIMRQE I GWFDVHDV ...文字列: MDLEGDRNGG AKKKNFFKLN NKSEKDKKEK KPTVSVFSMF RYSNWLDKLY MVVGTLAAII HGAGLPLMML VFGEMTDIFA NAGNLEDLM SNITNRSDIN DTGFFMNLEE DMTRYAYYYS GIGAGVLVAA YIQVSFWCLA AGRQIHKIRK QFFHAIMRQE I GWFDVHDV GELNTRLTDD VSKINEGIGD KIGMFFQSMA TFFTGFIVGF TRGWKLTLVI LAISPVLGLS AAVWAKILSS FT DKELLAY AKAGAVAEEV LAAIRTVIAF GGQKKELERY NKNLEEAKRI GIKKAITANI SIGAAFLLIY ASYALAFWYG TTL VLSGEY SIGQVLTVFF SVLIGAFSVG QASPSIEAFA NARGAAYEIF KIIDNKPSID SYSKSGHKPD NIKGNLEFRN VHFS YPSRK EVKILKGLNL KVQSGQTVAL VGNSGCGKST TVQLMQRLYD PTEGMVSVDG QDIRTINVRF LREIIGVVSQ EPVLF ATTI AENIRYGREN VTMDEIEKAV KEANAYDFIM KLPHKFDTLV GERGAQLSGG QKQRIAIARA LVRNPKILLL DQATSA LDT ESEAVVQVAL DKARKGRTTI VIAHRLSTVR NADVIAGFDD GVIVEKGNHD ELMKEKGIYF KLVTMQTAGN EVELENA AD ESKSEIDALE MSSNDSRSSL IRKRSTRRSV RGSQAQDRKL STKEALDESI PPVSFWRIMK LNLTEWPYFV VGVFCAII N GGLQPAFAII FSKIIGVFTR IDDPETKRQN SNLFSLLFLA LGIISFITFF LQGFTFGKAG EILTKRLRYM VFRSMLRQD VSWFDDPKNT TGALTTRLAN DAAQVKGAIG SRLAVITQNI ANLGTGIIIS FIYGWQLTLL LLAIVPIIAI AGVVEMKMLS GQALKDKKE LEGSGKIATE AIENFRTVVS LTQEQKFEHM YAQSLQVPYR NSLRKAHIFG ITFSFTQAMM YFSYAGCFRF G AYLVAHKL MSFEDVLLVF SAVVFGAMAV GQVSSFAPDY AKAKISAAHI IMIIEKTPLI DSYSTEGLMP NTLEGNVTFG EV VFNYPTR PDIPVLQGLS LEVKKGQTLA LVGSSGCGKS TVVQLLERFY DPLAGKVLLD GKEIKRLNVQ WLRAHLGIVS QEP ILFDCS IAENIAYGDN SRVVSQEEIV RAAKEANIHA FIESLPNKYS TKVGDKGTQL SGGQKQRIAI ARALVRQPHI LLLD QATSA LDTESEKVVQ EALDKAREGR TCIVIAHRLS TIQNADLIVV FQNGRVKEHG THQQLLAQKG IYFSMVSVQA GTKRQ SNSL EVLFQ UniProtKB: ATP-dependent translocase ABCB1 |
-分子 #2: ADENOSINE-5'-TRIPHOSPHATE
| 分子 | 名称: ADENOSINE-5'-TRIPHOSPHATE / タイプ: ligand / ID: 2 / コピー数: 2 / 式: ATP |
|---|---|
| 分子量 | 理論値: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-分子 #3: MAGNESIUM ION
| 分子 | 名称: MAGNESIUM ION / タイプ: ligand / ID: 3 / コピー数: 2 / 式: MG |
|---|---|
| 分子量 | 理論値: 24.305 Da |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 5.0 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.5 構成要素:
| ||||||||||||||||||
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: GOLD / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY ARRAY / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 15 sec. / 前処理 - 雰囲気: AIR | ||||||||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 295 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 温度 | 最低: 80.0 K / 最高: 100.0 K |
| 特殊光学系 | エネルギーフィルター - エネルギー下限: 10 eV エネルギーフィルター - エネルギー上限: 10 eV |
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: SUPER-RESOLUTION / デジタル化 - サイズ - 横: 3838 pixel / デジタル化 - サイズ - 縦: 3710 pixel / デジタル化 - 画像ごとのフレーム数: 1-50 / 撮影したグリッド数: 2 / 実像数: 5747 / 平均露光時間: 7.0 sec. / 平均電子線量: 80.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 70.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.2 µm / 最小 デフォーカス(公称値): 0.8 µm |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | 空間: RECIPROCAL / プロトコル: FLEXIBLE FIT |
|---|---|
| 得られたモデル |  PDB-6c0v: |
 ムービー
ムービー コントローラー
コントローラー
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)