+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7128 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the mechanically activated ion channel Piezo1 | |||||||||||||||
 Map data Map data | C3 symmetry refinement mouse Piezo1 core. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | mechanosensitive ion channel / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmechanosensitive monoatomic cation channel activity / cuticular plate / positive regulation of cell-cell adhesion mediated by integrin / positive regulation of integrin activation / detection of mechanical stimulus / mechanosensitive monoatomic ion channel activity / stereocilium / positive regulation of myotube differentiation / lamellipodium membrane / monoatomic cation transport ...mechanosensitive monoatomic cation channel activity / cuticular plate / positive regulation of cell-cell adhesion mediated by integrin / positive regulation of integrin activation / detection of mechanical stimulus / mechanosensitive monoatomic ion channel activity / stereocilium / positive regulation of myotube differentiation / lamellipodium membrane / monoatomic cation transport / monoatomic cation channel activity / endoplasmic reticulum-Golgi intermediate compartment membrane / regulation of membrane potential / endoplasmic reticulum membrane / endoplasmic reticulum / identical protein binding / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||||||||
 Authors Authors | Saotome K / Kefauver JM / Patapoutian A / Ward AB | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Structure of the mechanically activated ion channel Piezo1. Authors: Kei Saotome / Swetha E Murthy / Jennifer M Kefauver / Tess Whitwam / Ardem Patapoutian / Andrew B Ward /  Abstract: Piezo1 and Piezo2 are mechanically activated ion channels that mediate touch perception, proprioception and vascular development. Piezo proteins are distinct from other ion channels and their ...Piezo1 and Piezo2 are mechanically activated ion channels that mediate touch perception, proprioception and vascular development. Piezo proteins are distinct from other ion channels and their structure remains poorly defined, which impedes detailed study of their gating and ion permeation properties. Here we report a high-resolution cryo-electron microscopy structure of the mouse Piezo1 trimer. The detergent-solubilized complex adopts a three-bladed propeller shape with a curved transmembrane region containing at least 26 transmembrane helices per protomer. The flexible propeller blades can adopt distinct conformations, and consist of a series of four-transmembrane helical bundles that we term Piezo repeats. Carboxy-terminal domains line the central ion pore, and the channel is closed by constrictions in the cytosol. A kinked helical beam and anchor domain link the Piezo repeats to the pore, and are poised to control gating allosterically. The structure provides a foundation to dissect further how Piezo channels are regulated by mechanical force. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7128.map.gz emd_7128.map.gz | 226.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7128-v30.xml emd-7128-v30.xml emd-7128.xml emd-7128.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7128.png emd_7128.png | 36.3 KB | ||
| Filedesc metadata |  emd-7128.cif.gz emd-7128.cif.gz | 6.3 KB | ||
| Others |  emd_7128_additional.map.gz emd_7128_additional.map.gz | 206.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7128 http://ftp.pdbj.org/pub/emdb/structures/EMD-7128 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7128 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7128 | HTTPS FTP |
-Related structure data
| Related structure data |  6bpzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7128.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7128.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C3 symmetry refinement mouse Piezo1 core. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Refinement of symmetry-expanded mouse Piezo1 blade class 1.
| File | emd_7128_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement of symmetry-expanded mouse Piezo1 blade class 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
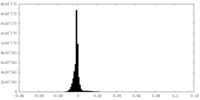
| Density Histograms |
- Sample components
Sample components
-Entire : mouse Piezo1
| Entire | Name: mouse Piezo1 |
|---|---|
| Components |
|
-Supramolecule #1: mouse Piezo1
| Supramolecule | Name: mouse Piezo1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 876 KDa |
-Macromolecule #1: Piezo-type mechanosensitive ion channel component 1,Piezo-type me...
| Macromolecule | Name: Piezo-type mechanosensitive ion channel component 1,Piezo-type mechanosensitive ion channel component 1,mouse Piezo1,Piezo-type mechanosensitive ion channel component 1,Piezo-type ...Name: Piezo-type mechanosensitive ion channel component 1,Piezo-type mechanosensitive ion channel component 1,mouse Piezo1,Piezo-type mechanosensitive ion channel component 1,Piezo-type mechanosensitive ion channel component 1 type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 161.973531 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PNPIPNFIHC RSYLDMLKVA VFRYLFWLVL VVVFVAGATR ISIFGLGYLL ACFYLLLFGT TLLQKDTRAQ LVLWDCLILY NVTVIISKN MLSLLSCVFV EQMQSNFCWV IQLFSLVCTV KGYYDPKEMM TRDRDCLLPV EEAGIIWDSI CFFFLLLQRR I FLSHYFLH ...String: PNPIPNFIHC RSYLDMLKVA VFRYLFWLVL VVVFVAGATR ISIFGLGYLL ACFYLLLFGT TLLQKDTRAQ LVLWDCLILY NVTVIISKN MLSLLSCVFV EQMQSNFCWV IQLFSLVCTV KGYYDPKEMM TRDRDCLLPV EEAGIIWDSI CFFFLLLQRR I FLSHYFLH VSADLKATAL QASRGFALYN AANLKSINFH RQIEEKSLAQ LKRQMKRIRA KQEKYRQSQA SRGQLQSKDP QD PSQEPGP DSPGGSSPPR RQWWRPWLDH ATVIHSGDYF LFESDSEEEE EALPEDPRPA AQSAFQMAYQ AWVTNAQTVL RQR RERARQ ERAEQLASGG DLNPDVEPVD VPEDEMAGRS HMMQRVLSTM QFLWVLGQAT VDGLTRWLRA FTKHHRTMSD VLCA ERYLL TQELLRVGEV RRGVLDQLYV GEDEATLSGP VETRDGPSTA SSGLGAEEPL SSMTDD(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)P ELEEAERFEA QQGRTLRLLR AGYQCVAAHS ELLCYFIII LNHMVTASAA SLVLPVLVFL WAMLTIPRPS KRFWMTAIVF TEVMVVTKYL FQFGFFPWNS YVVLRRYENK PYFPPRILGL EKTDSYIKY DLVQLMALFF HRSQLLCYGL WDHEEDRYPK DHCRSSVKDR EAKEEPEAKL ESQSETGTGH PKEPVLAGTP R DHIQGKGS IRSKDVIQDP PEDLKPRHTR HISIRFRRRK ETPGPKGTAV METEHEEGEG KETTERKRPR HTQEKSKFRE RM KAAGRRL QSFCVSLAQS FYQPLQRFFH DILHTKYRAA TDVYALMFLA DIVDIIIIIF GFWAFGKHSA ATDIASSLSD DQV PQAFLF MLLVQFGTMV IDRALYLRKT VLGKLAFQVV LVVAIHIWMF FILPAVTERM FSQNAVAQLW YFVKCIYFAL SAYQ IRCGY PTRILGNFLT KKYNHLNLFL FQGFRLVPFL VELRAVMDWV WTDTTLSLSN WMCVEDIYAN IFIIKCSRET EKKYP QPKG QKKKKIVKYG MGGLIILFLI AIIWFPLLFM SLIRSVVGVV NQPIDVTVTL KLGGYEPLFT MSAQQPSIVP FTPQAY EEL SQQFDPYPLA MQFISQYSPE DIVTAQIEGS SGALWRISPP SRAQMKQELY NGTADITLRF TWNFQRDLAK GGTVEYT NE KHTLELAPNS TARRQLAQLL EGRPDQSVVI PHLFPKYIRA PNGPEANPVK QLQPDEEEDY LGVRIQLRRE QVGTGASG E QAGTKASDFL EWWVIELQDC KADCNLLPMV IFSDKVSPPS LGFLAGYGIV GLYVSIVLVV GKFVRGFFSE ISHSIMFEE LPCVDRILKL CQDIFLVRET RELELEEELY AKLIFLYRSP ETMIKWTRER EKKLGAPLEV LFQ UniProtKB: Piezo-type mechanosensitive ion channel component 1, Piezo-type mechanosensitive ion channel component 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil, UltrAuFoil / Material: GOLD / Mesh: 200 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. |
| Vitrification | Cryogen name: ETHANE |
| Details | mouse Piezo1 was purified in glyco-diosgenin. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 72627 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: RELION (ver. 2.0) |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: RELION (ver. 2.0) |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6bpz: |
 Movie
Movie Controller
Controller





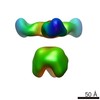
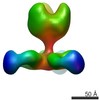




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)