+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Mycobacterium smegmatis EtfD | |||||||||
 Map data Map data | CryoEM map of Mycobacterium smegmatis EtfD (sharpened) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | oxidoreductase / membrane protein / electron transport chain / beta oxidation | |||||||||
| Function / homology |  Function and homology information Function and homology information4 iron, 4 sulfur cluster binding / oxidoreductase activity / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Courbon GM / Rubinstein JL | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Mycobacterial EtfD contains an unusual linear [3Fe-4S] cluster and enables beta oxidation to drive proton pumping by the electron transport chain Authors: Courbon GM / Rubinstein JL | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_70545.map.gz emd_70545.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-70545-v30.xml emd-70545-v30.xml emd-70545.xml emd-70545.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_70545_fsc.xml emd_70545_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_70545.png emd_70545.png | 68.2 KB | ||
| Filedesc metadata |  emd-70545.cif.gz emd-70545.cif.gz | 6.3 KB | ||
| Others |  emd_70545_additional_1.map.gz emd_70545_additional_1.map.gz emd_70545_half_map_1.map.gz emd_70545_half_map_1.map.gz emd_70545_half_map_2.map.gz emd_70545_half_map_2.map.gz | 63 MB 116.2 MB 116.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-70545 http://ftp.pdbj.org/pub/emdb/structures/EMD-70545 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-70545 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-70545 | HTTPS FTP |
-Related structure data
| Related structure data |  9ojnMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_70545.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_70545.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of Mycobacterium smegmatis EtfD (sharpened) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.024 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: CryoEM map of Mycobacterium smegmatis EtfD (unsharpened)
| File | emd_70545_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of Mycobacterium smegmatis EtfD (unsharpened) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoEM map of Mycobacterium smegmatis EtfD (Half B)
| File | emd_70545_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of Mycobacterium smegmatis EtfD (Half B) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoEM map of Mycobacterium smegmatis EtfD (Half A)
| File | emd_70545_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of Mycobacterium smegmatis EtfD (Half A) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of Mycobacterium smegmatis EtfD
| Entire | Name: Structure of Mycobacterium smegmatis EtfD |
|---|---|
| Components |
|
-Supramolecule #1: Structure of Mycobacterium smegmatis EtfD
| Supramolecule | Name: Structure of Mycobacterium smegmatis EtfD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
-Macromolecule #1: Iron-sulfur-binding reductase
| Macromolecule | Name: Iron-sulfur-binding reductase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 111.045625 KDa |
| Sequence | String: MAHTLEVSRL IIGLLMTAIV LVFAAKRVLW LTKLIRSGQK TLDENGRKND LQKRITTQIT EVFGQTRLLR WSVPGIAHFF TMWGFFVLA SVYLEAYGVL FDPEFHIPFI GRWPVLGFLQ DFFAVAVLLG IIVFAIIRVV REPKKIGRDS RFYGSHTGGA W EILFMIFL ...String: MAHTLEVSRL IIGLLMTAIV LVFAAKRVLW LTKLIRSGQK TLDENGRKND LQKRITTQIT EVFGQTRLLR WSVPGIAHFF TMWGFFVLA SVYLEAYGVL FDPEFHIPFI GRWPVLGFLQ DFFAVAVLLG IIVFAIIRVV REPKKIGRDS RFYGSHTGGA W EILFMIFL VIATYALFRG AAVNTLGERF PYQSGAFFSD FMAWILRPLG ATANMWIETV ALMGHIGVML VFLLIVLHSK HL HIGLAPI NVTFKRLPNG LGPLLPMESN GEYIDFEDPA EDAVFGKGKI EDFTWKGYLD FTTCTECGRC QSQCPAWNTG KPL SPKLVI MNLRDHMFAK APYILGEKPS PLESTPEGGL GEKARGEKHE QKHAHDHVPE SGFERILGSG PEQALRPLVG TEEQ GGVID PDVLWSCTNC GACVEQCPVD IEHIDHIVDM RRYQVMVESE FPGELGVLFK NLETKGNPWG QNAKDRTNWI DEVDF DVPV YGEDVDSFDG FEYLFWVGCA GAYEDRAKKT TKAVAELLAT AGVKFLVLGT GETCTGDSAR RSGNEFLFQQ LAAQNV ETI NELFEGVETV DRKIVVTCPH CFNTIGREYP QLGANYSVVH HTQLLNRLVR DKKLVPVKSV SEQNGQPVTY HDPCFLG RH NKVYEAPREL VEASGVTLKE MPRHADRGLC CGAGGARMWM EEHIGKRVNV ERTEEAMDTA STIATGCPFC RVMITDGV D DVAASRNVEK AEVLDVAQLL LNSLDTSKVT LPEKGTAAKE SEKRAAARAE AEAKAEAAAP PVEEAAPEAE APAAPAAGG AEAKPVTGLG MAGAAKRPGA KKAAPAAEAS AAPAAAPAPA KGLGLAGGAK RPGAKKAAAP AAEAPAAPAS DAPPVKGLGL AGGAKRPGA KKTAAAAPAE KPAATEAPEA SATPAAPAAP VKGLGLAAGA KRPGAKKTAA APAEKPAAAE TEAPAPAETA A PAEPAKPE PPVVGLGIAA GARRPGAKKA AAKPAAAPAP AAEKPAEQAA EPEKPAEKPA EPEKPEPPVV GLGIKPGAKR PG KR UniProtKB: Iron-sulfur-binding reductase |
-Macromolecule #2: DODECYL-BETA-D-MALTOSIDE
| Macromolecule | Name: DODECYL-BETA-D-MALTOSIDE / type: ligand / ID: 2 / Number of copies: 1 / Formula: LMT |
|---|---|
| Molecular weight | Theoretical: 510.615 Da |
| Chemical component information |  ChemComp-LMT: |
-Macromolecule #3: MENAQUINONE-9
| Macromolecule | Name: MENAQUINONE-9 / type: ligand / ID: 3 / Number of copies: 1 / Formula: MQ9 |
|---|---|
| Molecular weight | Theoretical: 785.233 Da |
| Chemical component information |  ChemComp-MQ9: |
-Macromolecule #4: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 4 / Number of copies: 1 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 2 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #6: Non-cubane [4Fe-4S]-cluster
| Macromolecule | Name: Non-cubane [4Fe-4S]-cluster / type: ligand / ID: 6 / Number of copies: 1 / Formula: 9S8 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information | 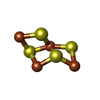 ChemComp-9S8: |
-Macromolecule #7: 1,3,5,7-tetrathia-2$l^{2},4$l^{4},6$l^{2}-triferraspiro[3.3]heptane
| Macromolecule | Name: 1,3,5,7-tetrathia-2$l^{2},4$l^{4},6$l^{2}-triferraspiro[3.3]heptane type: ligand / ID: 7 / Number of copies: 1 / Formula: A1CBX |
|---|---|
| Molecular weight | Theoretical: 295.795 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































