[English] 日本語
 Yorodumi
Yorodumi- EMDB-70510: Cryo-EM Structure of the Arabidopsis GA3-GID1A-RGA-SLY1-ASK1 Comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the Arabidopsis GA3-GID1A-RGA-SLY1-ASK1 Complex (Alternative Conformation) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RGA / DELLA / GID1 / SLY1 / ASK1 / GA / PLANT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of gibberellic acid mediated signaling pathway / fruit morphogenesis / gibberellin mediated signaling pathway / positive regulation of gibberellic acid mediated signaling pathway / raffinose family oligosaccharide biosynthetic process / floral organ morphogenesis / negative regulation of trichome patterning / negative regulation of developmental vegetative growth / negative regulation of leaf development / regulation of seed dormancy process ...regulation of gibberellic acid mediated signaling pathway / fruit morphogenesis / gibberellin mediated signaling pathway / positive regulation of gibberellic acid mediated signaling pathway / raffinose family oligosaccharide biosynthetic process / floral organ morphogenesis / negative regulation of trichome patterning / negative regulation of developmental vegetative growth / negative regulation of leaf development / regulation of seed dormancy process / gibberellin binding / seed dormancy process / negative regulation of gibberellic acid mediated signaling pathway / positive regulation of fertilization / meiotic cytokinesis / response to gibberellin / gibberellic acid mediated signaling pathway / regulation of seed germination / phragmoplast / jasmonic acid mediated signaling pathway / ethylene-activated signaling pathway / seed germination / response to far red light / Hydrolases / response to jasmonic acid / response to auxin / auxin-activated signaling pathway / negative regulation of DNA recombination / SCF ubiquitin ligase complex / regulation of protein catabolic process / response to cold / chromosome segregation / promoter-specific chromatin binding / microtubule cytoskeleton organization / spindle / cellular response to hypoxia / ubiquitin-dependent protein catabolic process / transcription cis-regulatory region binding / protein ubiquitination / DNA-binding transcription factor activity / hydrolase activity / positive regulation of gene expression / regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / mitochondrion / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.81 Å | |||||||||
 Authors Authors | Dahal P / Sharma K / Borgnia M / Zhou P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: Structural insights into proteolysis-dependent and -independent suppression of the master regulator DELLA by the gibberellin receptor. Authors: Pawan Dahal / Yan Wang / Jianhong Hu / Jeongmoo Park / Karly Forker / Zhong-Lin Zhang / Kedar Sharma / Mario J Borgnia / Tai-Ping Sun / Pei Zhou /  Abstract: The perception of the phytohormone gibberellin (GA) by its nuclear receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) triggers polyubiquitination and proteasomal degradation of master growth regulators- ...The perception of the phytohormone gibberellin (GA) by its nuclear receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) triggers polyubiquitination and proteasomal degradation of master growth regulators-DELLA proteins-mediated by the SCF E3 ubiquitin ligase complex. DELLA-encoding genes are known as 'Green Revolution' genes, as their dominant mutations lead to semidwarf cereal varieties with significantly higher yields due to reduced GA response. DELLAs function as central signaling hubs, coordinating diverse physiological responses by interacting with key transcription factors across multiple cellular pathways. While the DELLA domain mediates GA-GID1 binding, the mechanism of SCF recruitment remained unknown. Additionally, GA-GID1 binding can inhibit DELLA protein activity independently of its proteolysis, although the underlying mechanism was unclear. Here, we present the cryo-EM structures of GA-GID1A complexed with a full-length DELLA protein in , RGA (REPRESSOR OF ), and the GA-GID1A-RGA-SLY1-ASK1 complex. We show that the DELLA domain of RGA functions as a molecular bridge to enhance its GRAS domain binding to GID1A through direct interactions with both the GRAS domain and GID1A. Disrupting either intramolecular (DELLA-GRAS) or intermolecular (GRAS-GID1A) interactions weakens RGA-GID1 binding. Contrary to prior models, SLY1 binds the GRAS domain's concave surface without inducing conformational changes. Combining AlphaFold modeling and yeast three-hybrid assays, we demonstrate that GID1 binding to the RGA GRAS domain blocks its interactions with INDETERMINATE DOMAIN (IDD) transcription factors, explaining how GA-GID1 relieves growth suppression independently of DELLA degradation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_70510.map.gz emd_70510.map.gz | 59.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-70510-v30.xml emd-70510-v30.xml emd-70510.xml emd-70510.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_70510_fsc.xml emd_70510_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_70510.png emd_70510.png | 115.3 KB | ||
| Masks |  emd_70510_msk_1.map emd_70510_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-70510.cif.gz emd-70510.cif.gz | 6.9 KB | ||
| Others |  emd_70510_additional_1.map.gz emd_70510_additional_1.map.gz emd_70510_half_map_1.map.gz emd_70510_half_map_1.map.gz emd_70510_half_map_2.map.gz emd_70510_half_map_2.map.gz | 2 MB 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-70510 http://ftp.pdbj.org/pub/emdb/structures/EMD-70510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-70510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-70510 | HTTPS FTP |
-Related structure data
| Related structure data |  9oi8MC  9o4jC  9o4kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_70510.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_70510.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8469 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_70510_msk_1.map emd_70510_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_70510_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_70510_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_70510_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GA3-GID1A-RGA-SLY1-ASK1 Complex
| Entire | Name: GA3-GID1A-RGA-SLY1-ASK1 Complex |
|---|---|
| Components |
|
-Supramolecule #1: GA3-GID1A-RGA-SLY1-ASK1 Complex
| Supramolecule | Name: GA3-GID1A-RGA-SLY1-ASK1 Complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: DELLA protein RGA
| Macromolecule | Name: DELLA protein RGA / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65.253309 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKRDHHQFQG RLSNHGTSSS SSSISKDKMM MVKKEEDGGG NMDDELLAVL GYKVRSSEMA EVALKLEQLE TMMSNVQEDG LSHLATDTV HYNPSELYSW LDNMLSELNP PPLPASSNGL DPVLPSPEIC GFPASDYDLK VIPGNAIYQF PAIDSSSSSN N QNQQLQSC ...String: MKRDHHQFQG RLSNHGTSSS SSSISKDKMM MVKKEEDGGG NMDDELLAVL GYKVRSSEMA EVALKLEQLE TMMSNVQEDG LSHLATDTV HYNPSELYSW LDNMLSELNP PPLPASSNGL DPVLPSPEIC GFPASDYDLK VIPGNAIYQF PAIDSSSSSN N QNQQLQSC SSPDSMVTST STGTQIGGVI GTTVTTTTTT TTAAGESTRS VILVDSQENG VRLVHALMAC AEAIQQNNLT LA EALVKQI GCLAVSQAGA MRKVATYFAE ALARRIYRLS PPQNQIDHCL SDTLQMHFYE TCPYLKFAHF TANQAILEAF EGK KRVHVI DFSMNQGLQW PALMQALALR EGGPPTFRLT GIGPPAPDNS DHLHEVGCKL AQLAEAIHVE FEYRGFVANS LADL DASML ELRPSDTEAV AVNSVFELHK LLGRPGGIEK VLGVVKQIKP VIFTVVEQES NHNGPVFLDR FTESLHYYST LFDSL EGVP NSQDKVMSEV YLGKQICNLV ACEGPDRVER HETLSQWGNR FGSSGLAPAH LGSNAFKQAS MLLSVFNSGQ GYRVEE SNG CLMLGWHTRP LITTSAWKLS TAAYGGWSHP QFER UniProtKB: DELLA protein RGA |
-Macromolecule #2: Gibberellin receptor GID1A
| Macromolecule | Name: Gibberellin receptor GID1A / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.775859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAASDEVNLI ESRTVVPLNT WVLISNFKVA YNILRRPDGT FNRHLAEYLD RKVTANANPV DGVFSFDVLI DRRINLLSRV YRPAYADQE QPPSILDLEK PVDGDIVPVI LFFHGGSFAH SSANSAIYDT LCRRLVGLCK CVVVSVNYRR APENPYPCAY D DGWIALNW ...String: MAASDEVNLI ESRTVVPLNT WVLISNFKVA YNILRRPDGT FNRHLAEYLD RKVTANANPV DGVFSFDVLI DRRINLLSRV YRPAYADQE QPPSILDLEK PVDGDIVPVI LFFHGGSFAH SSANSAIYDT LCRRLVGLCK CVVVSVNYRR APENPYPCAY D DGWIALNW VNSRSWLKSK KDSKVHIFLA GDSSGGNIAH NVALRAGESG IDVLGNILLN PMFGGNERTE SEKSLDGKYF VT VRDRDWY WKAFLPEGED REHPACNPFS PRGKSLEGVS FPKSLVVVAG LDLIRDWQLA YAEGLKKAGQ EVKLMHLEKA TVG FYLLPN NNHFHNVMDE ISAFVNAECG GDYKDDDDK UniProtKB: Gibberellin receptor GID1A |
-Macromolecule #3: F-box protein GID2
| Macromolecule | Name: F-box protein GID2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.069963 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHHHHH ENLYFQSHMK RSTTDSDLAG DAHNETNKKM KSTEEEEIGF SNLDENLVYE VLKHVDAKTL AMSSCVSKIW HKTAQDERL WELICTRHWT NIGCGQNQLR SVVLALGGFR RLHSLYLWPL SKPNPRARFG KDELKLTLSL LSIRYYKKMS F TKRPLPES KGGWSHPQFE R UniProtKB: F-box protein GID2 |
-Macromolecule #4: SKP1-like protein 1A
| Macromolecule | Name: SKP1-like protein 1A / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.273586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHHHHH ENLYFQSHMS AKKIVLKSSD GESFEVEEAV ALESQTIAHM VEDDCVDNGV PLPNVTSKIL AKVIEYCKRH VEAAASKAE AVEGAATSDD DLKAWDADFM KIDQATLFEL ILAANYLNIK NLLDLTCQTV ADMIKGKTPE EIRTTFNIKN D FTPEEEEE VRRENQWAFE GGYKDDDDK UniProtKB: SKP1-like protein 1A |
-Macromolecule #5: GIBBERELLIN A3
| Macromolecule | Name: GIBBERELLIN A3 / type: ligand / ID: 5 / Number of copies: 1 / Formula: GA3 |
|---|---|
| Molecular weight | Theoretical: 346.374 Da |
| Chemical component information | 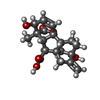 ChemComp-GA3: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















































