[English] 日本語
 Yorodumi
Yorodumi- EMDB-7033: Emptied phiX174 complexed with lipopolysaccharides after DNA ejection -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7033 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Emptied phiX174 complexed with lipopolysaccharides after DNA ejection | |||||||||
 Map data Map data | Emptied phiX174 complexed with lipopolysaccharides after DNA ejection, low pass-filtered to 8 Angstrom resolution | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Enterobacteria phage phiX174 isolate Sanger (virus) / Enterobacteria phage phiX174 isolate Sanger (virus) /  Salmonella enterica (bacteria) Salmonella enterica (bacteria) | |||||||||
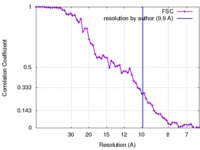
| Method | single particle reconstruction / cryo EM / Resolution: 9.9 Å | |||||||||
 Authors Authors | Rossmann MG / Sun Y / Klose T / Roznowski A / Fane BA / Pollack L / Tokuda J / Mauney A | |||||||||
 Citation Citation |  Journal: Nature / Year: 2014 Journal: Nature / Year: 2014Title: Icosahedral bacteriophage ΦX174 forms a tail for DNA transport during infection. Authors: Lei Sun / Lindsey N Young / Xinzheng Zhang / Sergei P Boudko / Andrei Fokine / Erica Zbornik / Aaron P Roznowski / Ian J Molineux / Michael G Rossmann / Bentley A Fane /  Abstract: Prokaryotic viruses have evolved various mechanisms to transport their genomes across bacterial cell walls. Many bacteriophages use a tail to perform this function, whereas tail-less phages rely on ...Prokaryotic viruses have evolved various mechanisms to transport their genomes across bacterial cell walls. Many bacteriophages use a tail to perform this function, whereas tail-less phages rely on host organelles. However, the tail-less, icosahedral, single-stranded DNA ΦX174-like coliphages do not fall into these well-defined infection processes. For these phages, DNA delivery requires a DNA pilot protein. Here we show that the ΦX174 pilot protein H oligomerizes to form a tube whose function is most probably to deliver the DNA genome across the host's periplasmic space to the cytoplasm. The 2.4 Å resolution crystal structure of the in vitro assembled H protein's central domain consists of a 170 Å-long α-helical barrel. The tube is constructed of ten α-helices with their amino termini arrayed in a right-handed super-helical coiled-coil and their carboxy termini arrayed in a left-handed super-helical coiled-coil. Genetic and biochemical studies demonstrate that the tube is essential for infectivity but does not affect in vivo virus assembly. Cryo-electron tomograms show that tubes span the periplasmic space and are present while the genome is being delivered into the host cell's cytoplasm. Both ends of the H protein contain transmembrane domains, which anchor the assembled tubes into the inner and outer cell membranes. The central channel of the H-protein tube is lined with amide and guanidinium side chains. This may be a general property of viral DNA conduits and is likely to be critical for efficient genome translocation into the host. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7033.map.gz emd_7033.map.gz | 13.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7033-v30.xml emd-7033-v30.xml emd-7033.xml emd-7033.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7033_fsc.xml emd_7033_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_7033.png emd_7033.png | 177 KB | ||
| Masks |  emd_7033_msk_1.map emd_7033_msk_1.map emd_7033_msk_2.map emd_7033_msk_2.map | 15.6 MB 15.6 MB |  Mask map Mask map | |
| Others |  emd_7033_half_map_1.map.gz emd_7033_half_map_1.map.gz emd_7033_half_map_2.map.gz emd_7033_half_map_2.map.gz | 11.9 MB 11.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7033 http://ftp.pdbj.org/pub/emdb/structures/EMD-7033 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7033 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7033 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7033.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7033.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Emptied phiX174 complexed with lipopolysaccharides after DNA ejection, low pass-filtered to 8 Angstrom resolution | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7033_msk_1.map emd_7033_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_7033_msk_2.map emd_7033_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Emptied phiX174 complexed with lipopolysaccharides after DNA ejection,...
| File | emd_7033_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Emptied phiX174 complexed with lipopolysaccharides after DNA ejection, half map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Emptied phiX174 complexed with lipopolysaccharides after DNA ejection,...
| File | emd_7033_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Emptied phiX174 complexed with lipopolysaccharides after DNA ejection, half map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Full phiX174 complexed with lipopolysaccharides before DNA ejection
| Entire | Name: Full phiX174 complexed with lipopolysaccharides before DNA ejection |
|---|---|
| Components |
|
-Supramolecule #1: Full phiX174 complexed with lipopolysaccharides before DNA ejection
| Supramolecule | Name: Full phiX174 complexed with lipopolysaccharides before DNA ejection type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage phiX174 isolate Sanger (virus) Enterobacteria phage phiX174 isolate Sanger (virus) |
| Molecular weight | Theoretical: 10 KDa |
-Supramolecule #2: Lipopolysaccharides (rough strains) from Salmonella enterica sero...
| Supramolecule | Name: Lipopolysaccharides (rough strains) from Salmonella enterica serotype typhimurium TV119 (Ra mutant) type: organelle_or_cellular_component / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Salmonella enterica (bacteria) / Strain: typhimurium TV119 / Location in cell: outer membrane Salmonella enterica (bacteria) / Strain: typhimurium TV119 / Location in cell: outer membrane |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 |
| Details | Mixed with LPS and incubated at 33 degrees C for 1 minute |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)