[English] 日本語
 Yorodumi
Yorodumi- EMDB-7021: Structure of influenza hemagglutinin from A/New Caledonia/20/99 i... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7021 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of influenza hemagglutinin from A/New Caledonia/20/99 in complex with FAB 441D6. | |||||||||
 Map data Map data | Hemagglutinin ectodomain derived from influenza A/New Caledonia/20/99 expressed with a trimerization domain (foldon), in complex with 3 molecules of FAB 441D6. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Influenza A virus Influenza A virus | |||||||||
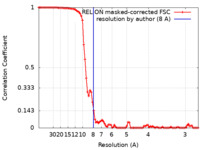
| Method | single particle reconstruction / cryo EM / Resolution: 8.0 Å | |||||||||
 Authors Authors | Gallagher JR / Harris AK | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Immunol / Year: 2019 Journal: Nat Immunol / Year: 2019Title: Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Authors: Masaru Kanekiyo / M Gordon Joyce / Rebecca A Gillespie / John R Gallagher / Sarah F Andrews / Hadi M Yassine / Adam K Wheatley / Brian E Fisher / David R Ambrozak / Adrian Creanga / Kwanyee ...Authors: Masaru Kanekiyo / M Gordon Joyce / Rebecca A Gillespie / John R Gallagher / Sarah F Andrews / Hadi M Yassine / Adam K Wheatley / Brian E Fisher / David R Ambrozak / Adrian Creanga / Kwanyee Leung / Eun Sung Yang / Seyhan Boyoglu-Barnum / Ivelin S Georgiev / Yaroslav Tsybovsky / Madhu S Prabhakaran / Hanne Andersen / Wing-Pui Kong / Ulrich Baxa / Kathryn L Zephir / Julie E Ledgerwood / Richard A Koup / Peter D Kwong / Audray K Harris / Adrian B McDermott / John R Mascola / Barney S Graham /   Abstract: The present vaccine against influenza virus has the inevitable risk of antigenic discordance between the vaccine and the circulating strains, which diminishes vaccine efficacy. This necessitates new ...The present vaccine against influenza virus has the inevitable risk of antigenic discordance between the vaccine and the circulating strains, which diminishes vaccine efficacy. This necessitates new approaches that provide broader protection against influenza. Here we designed a vaccine using the hypervariable receptor-binding domain (RBD) of viral hemagglutinin displayed on a nanoparticle (np) able to elicit antibody responses that neutralize H1N1 influenza viruses spanning over 90 years. Co-display of RBDs from multiple strains across time, so that the adjacent RBDs are heterotypic, provides an avidity advantage to cross-reactive B cells. Immunization with the mosaic RBD-np elicited broader antibody responses than those induced by an admixture of nanoparticles encompassing the same set of RBDs as separate homotypic arrays. Furthermore, we identified a broadly neutralizing monoclonal antibody in a mouse immunized with mosaic RBD-np. The mosaic antigen array signifies a unique approach that subverts monotypic immunodominance and allows otherwise subdominant cross-reactive B cell responses to emerge. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7021.map.gz emd_7021.map.gz | 62.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7021-v30.xml emd-7021-v30.xml emd-7021.xml emd-7021.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7021_fsc.xml emd_7021_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_7021.png emd_7021.png | 94.4 KB | ||
| Masks |  emd_7021_msk_1.map emd_7021_msk_1.map | 67 MB |  Mask map Mask map | |
| Others |  emd_7021_half_map_1.map.gz emd_7021_half_map_1.map.gz emd_7021_half_map_2.map.gz emd_7021_half_map_2.map.gz | 51.9 MB 51.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7021 http://ftp.pdbj.org/pub/emdb/structures/EMD-7021 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7021 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7021 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7021.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7021.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hemagglutinin ectodomain derived from influenza A/New Caledonia/20/99 expressed with a trimerization domain (foldon), in complex with 3 molecules of FAB 441D6. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3797 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
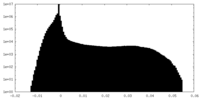
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7021_msk_1.map emd_7021_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
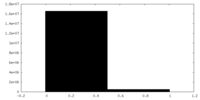
| Density Histograms |
-Half map: RELION unfiltered half map (1)
| File | emd_7021_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION unfiltered half map (1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
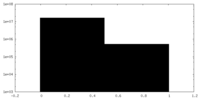
| Density Histograms |
-Half map: RELION unfiltered half map (2)
| File | emd_7021_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION unfiltered half map (2) | ||||||||||||
| Projections & Slices |
| ||||||||||||
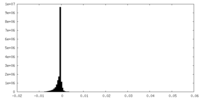
| Density Histograms |
- Sample components
Sample components
-Entire : Influenza hemagglutinin from A/New Caledonia/20/99, in complex wi...
| Entire | Name: Influenza hemagglutinin from A/New Caledonia/20/99, in complex with 3 molecules of FAB 441D6. |
|---|---|
| Components |
|
-Supramolecule #1: Influenza hemagglutinin from A/New Caledonia/20/99, in complex wi...
| Supramolecule | Name: Influenza hemagglutinin from A/New Caledonia/20/99, in complex with 3 molecules of FAB 441D6. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Influenza A virus / Strain: A/New Caledonia/20/99 Influenza A virus / Strain: A/New Caledonia/20/99 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Influenza hemagglutinin from A/New Caledonia/20/99, with C-termin...
| Macromolecule | Name: Influenza hemagglutinin from A/New Caledonia/20/99, with C-terminal trimerization domain (T4 fibritin foldon). type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus / Strain: A/New Caledonia/20/99 Influenza A virus / Strain: A/New Caledonia/20/99 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DTICIGYHAN NSTDTVDTVL EKNVTVTHSV NLLEDSHNGK LCRLKGTAPL QLGNCSVAGW ILGNPECESL FSKESWSYIA ETPNPENGTC YPGYFADYEE LREQLSSVSS FERFEIFPKE SSWPNHTVTK GVTASCSHNG KSSFYKNLLW LTEKNGLYPN LSKSYVNNKE ...String: DTICIGYHAN NSTDTVDTVL EKNVTVTHSV NLLEDSHNGK LCRLKGTAPL QLGNCSVAGW ILGNPECESL FSKESWSYIA ETPNPENGTC YPGYFADYEE LREQLSSVSS FERFEIFPKE SSWPNHTVTK GVTASCSHNG KSSFYKNLLW LTEKNGLYPN LSKSYVNNKE KEVLVLWGVH HPSNIGDQRA IYHTENAYVS VVSSHYSRRF TPEITKRPKV RDQEGRINYY WTLLEPGDTI IFEANGNLIA PWYAFALSRG FGSGIITSNA SMGECDAKCQ TPQGAINSSL PFQNVHPVTI GECPKYVRST KLRMVTGLRN IPSIQSRGLF GAIAGFIEGG WTGMIDGWYG YHHQNEQGSG YAADQKSTQN AIDGITNKVN SVIEKMNTQF TAVGKEFNKL ERRMENLNKK VDDGFLDIWT YNAELLVLLE NERTLDFHDS NVKNLYEKVK TQLKNNAKEI GNGCFEFYHK CNNECMESVK NGTYDYPKYS EESKLNREKI DGSGLVPRGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGSA HIVMVDAYKP TKGHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: PBS | |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | Complexes of hemagglutinin and FAB were purified from unbound components by gel filtration. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-7 / Number grids imaged: 1 / Number real images: 464 / Average exposure time: 1.0 sec. / Average electron dose: 34.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 8.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)