[English] 日本語
 Yorodumi
Yorodumi- EMDB-7006: Human ribonucleotide reductase large subunit (alpha) with dATP and CDP -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7006 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human ribonucleotide reductase large subunit (alpha) with dATP and CDP | ||||||||||||
 Map data Map data | Human ribonucleotide reductase large subunit (alpha) with dATP and CDP | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Ribonucleotide Reductase Electron transfer Radical chemistry Thiyl radical / OXIDOREDUCTASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationribonucleoside-diphosphate reductase activity / pyrimidine nucleobase metabolic process / cell proliferation in forebrain / ribonucleoside diphosphate metabolic process / 2'-deoxyribonucleotide biosynthetic process / positive regulation of G0 to G1 transition / mitochondrial DNA replication / ribonucleoside-diphosphate reductase complex / Interconversion of nucleotide di- and triphosphates / ribonucleoside-diphosphate reductase ...ribonucleoside-diphosphate reductase activity / pyrimidine nucleobase metabolic process / cell proliferation in forebrain / ribonucleoside diphosphate metabolic process / 2'-deoxyribonucleotide biosynthetic process / positive regulation of G0 to G1 transition / mitochondrial DNA replication / ribonucleoside-diphosphate reductase complex / Interconversion of nucleotide di- and triphosphates / ribonucleoside-diphosphate reductase / ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor / deoxyribonucleotide biosynthetic process / protein heterotetramerization / DNA synthesis involved in DNA repair / response to ionizing radiation / positive regulation of G2/M transition of mitotic cell cycle / positive regulation of G1/S transition of mitotic cell cycle / male gonad development / centriolar satellite / disordered domain specific binding / nuclear envelope / retina development in camera-type eye / ciliary basal body / DNA repair / neuronal cell body / mitochondrion / ATP binding / identical protein binding / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||
 Authors Authors | Brignole EJ / Drennan CL | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: 3.3-Å resolution cryo-EM structure of human ribonucleotide reductase with substrate and allosteric regulators bound. Authors: Edward J Brignole / Kuang-Lei Tsai / Johnathan Chittuluru / Haoran Li / Yimon Aye / Pawel A Penczek / JoAnne Stubbe / Catherine L Drennan / Francisco Asturias /  Abstract: Ribonucleotide reductases (RNRs) convert ribonucleotides into deoxyribonucleotides, a reaction essential for DNA replication and repair. Human RNR requires two subunits for activity, the α subunit ...Ribonucleotide reductases (RNRs) convert ribonucleotides into deoxyribonucleotides, a reaction essential for DNA replication and repair. Human RNR requires two subunits for activity, the α subunit contains the active site, and the β subunit houses the radical cofactor. Here, we present a 3.3-Å resolution structure by cryo-electron microscopy (EM) of a dATP-inhibited state of human RNR. This structure, which was determined in the presence of substrate CDP and allosteric regulators ATP and dATP, has three α units arranged in an α ring. At near-atomic resolution, these data provide insight into the molecular basis for CDP recognition by allosteric specificity effectors dATP/ATP. Additionally, we present lower-resolution EM structures of human α in the presence of both the anticancer drug clofarabine triphosphate and β. Together, these structures support a model for RNR inhibition in which β is excluded from binding in a radical transfer competent position when α exists as a stable hexamer. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7006.map.gz emd_7006.map.gz | 6.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7006-v30.xml emd-7006-v30.xml emd-7006.xml emd-7006.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7006.png emd_7006.png | 189.3 KB | ||
| Filedesc metadata |  emd-7006.cif.gz emd-7006.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7006 http://ftp.pdbj.org/pub/emdb/structures/EMD-7006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7006 | HTTPS FTP |
-Related structure data
| Related structure data |  6auiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7006.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7006.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human ribonucleotide reductase large subunit (alpha) with dATP and CDP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human ribonucleotide reductase large subnunit (alpha)
| Entire | Name: Human ribonucleotide reductase large subnunit (alpha) |
|---|---|
| Components |
|
-Supramolecule #1: Human ribonucleotide reductase large subnunit (alpha)
| Supramolecule | Name: Human ribonucleotide reductase large subnunit (alpha) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Ribonucleoside-diphosphate reductase large subunit
| Macromolecule | Name: Ribonucleoside-diphosphate reductase large subunit / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: ribonucleoside-diphosphate reductase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 92.350391 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MHVIKRDGRQ ERVMFDKITS RIQKLCYGLN MDFVDPAQIT MKVIQGLYSG VTTVELDTLA AETAATLTT KHPDYAILAA RIAVSNLHKE TKKVFSDVME DLYNYINPHN GKHSPMVAKS TLDIVLANKD RLNSAIIYDR D FSYNYFGF ...String: MGSSHHHHHH SSGLVPRGSH MHVIKRDGRQ ERVMFDKITS RIQKLCYGLN MDFVDPAQIT MKVIQGLYSG VTTVELDTLA AETAATLTT KHPDYAILAA RIAVSNLHKE TKKVFSDVME DLYNYINPHN GKHSPMVAKS TLDIVLANKD RLNSAIIYDR D FSYNYFGF KTLERSYLLK INGKVAERPQ HMLMRVSVGI HKEDIDAAIE TYNLLSERWF THASPTLFNA GTNRPQLSSC FL LSMKDDS IEGIYDTLKQ CALISKSAGG IGVAVSCIRA TGSYIAGTNG NSNGLVPMLR VYNNTARYVD QGGNKRPGAF AIY LEPWHL DIFEFLDLKK NTGKEEQRAR DLFFALWIPD LFMKRVETNQ DWSLMCPNEC PGLDEVWGEE FEKLYASYEK QGRV RKVVK AQQLWYAIIE SQTETGTPYM LYKDSCNRKS NQQNLGTIKC SNLCTEIVEY TSKDEVAVCN LASLALNMYV TSEHT YDFK KLAEVTKVVV RNLNKIIDIN YYPVPEACLS NKRHRPIGIG VQGLADAFIL MRYPFESAEA QLLNKQIFET IYYGAL EAS CDLAKEQGPY ETYEGSPVSK GILQYDMWNV TPTDLWDWKV LKEKIAKYGI RNSLLIAPMP TASTAQILGN NESIEPY TS NIYTRRVLSG EFQIVNPHLL KDLTERGLWH EEMKNQIIAC NGSIQSIPEI PDDLKQLYKT VWEISQKTVL KMAAERGA F IDQSQSLNIH IAEPNYGKLT SMHFYGWKQG LKTGMYYLRT RPAANPIQFT LNKEKLKDKE KVSKEEEEKE RNTAAMVCS LENRDECLMC GS UniProtKB: Ribonucleoside-diphosphate reductase large subunit |
-Macromolecule #2: 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE
| Macromolecule | Name: 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 12 / Formula: DTP |
|---|---|
| Molecular weight | Theoretical: 491.182 Da |
| Chemical component information |  ChemComp-DTP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 12 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: CYTIDINE-5'-DIPHOSPHATE
| Macromolecule | Name: CYTIDINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 6 / Formula: CDP |
|---|---|
| Molecular weight | Theoretical: 403.176 Da |
| Chemical component information | 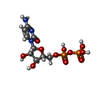 ChemComp-CDP: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 36 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||||||||
| Grid | Model: Protochips C-Flat / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER Details: glow discharged at 20 mA in an EMITech K100X Grid was first cleaned in a Solarus 950 (Gatan) at 25 W for 10 s in 75/25 Ar/O2 gas mixture | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 75 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER Details: manual blot with a strip of Whatman paper until drop stops wicking determined visually. | |||||||||||||||
| Details | 14 microM alpha and 0.05 mM dATP, 3 mM ATP, 1 mM CDP in 50 mM HEPES, pH 7.6, 15 mM MgCl2, 1 mM EDTA, 5 mM DTT, and 50 mM KCl |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 5-38 / Number real images: 2144 / Average exposure time: 7.6 sec. / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL / In silico model: Calculated ab initio |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D3 (2x3 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: SPARX / Number images used: 43885 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: SPARX |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6aui: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















