+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6999 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of T=1 Penaeus vannamei nodavirus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nodaviridae / shrimp nodavirus / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Icosahedral viral capsid protein, S domain / Viral coat protein (S domain) / T=3 icosahedral viral capsid / Viral coat protein subunit / structural molecule activity / metal ion binding / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Penaeus vannamei nodavirus Penaeus vannamei nodavirus | |||||||||
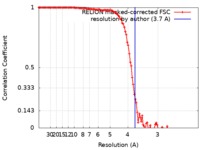
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Chen NC / Miyazaki N | |||||||||
| Funding support |  Taiwan, 1 items Taiwan, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2019 Journal: Commun Biol / Year: 2019Title: The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism. Authors: Nai-Chi Chen / Masato Yoshimura / Naoyuki Miyazaki / Hong-Hsiang Guan / Phimonphan Chuankhayan / Chien-Chih Lin / Shao-Kang Chen / Pei-Ju Lin / Yen-Chieh Huang / Kenji Iwasaki / Atsushi ...Authors: Nai-Chi Chen / Masato Yoshimura / Naoyuki Miyazaki / Hong-Hsiang Guan / Phimonphan Chuankhayan / Chien-Chih Lin / Shao-Kang Chen / Pei-Ju Lin / Yen-Chieh Huang / Kenji Iwasaki / Atsushi Nakagawa / Sunney I Chan / Chun-Jung Chen /    Abstract: Shrimp nodaviruses, including (PvNV) and nodaviruses (MrNV), cause white-tail disease in shrimps, with high mortality. The viral capsid structure determines viral assembly and host specificity ...Shrimp nodaviruses, including (PvNV) and nodaviruses (MrNV), cause white-tail disease in shrimps, with high mortality. The viral capsid structure determines viral assembly and host specificity during infections. Here, we show cryo-EM structures of = 3 and = 1 PvNV-like particles (PvNV-LPs), crystal structures of the protrusion-domains (P-domains) of PvNV and MrNV, and the crystal structure of the ∆N-ARM-PvNV shell-domain (S-domain) in = 1 subviral particles. The capsid protein of PvNV reveals five domains: the P-domain with a new jelly-roll structure forming cuboid-like spikes; the jelly-roll S-domain with two calcium ions; the linker between the S- and P-domains exhibiting new cross and parallel conformations; the N-arm interacting with nucleotides organized along icosahedral two-fold axes; and a disordered region comprising the basic -terminal arginine-rich motif (N-ARM) interacting with RNA. The N-ARM controls = 3 and = 1 assemblies. Increasing the /-termini flexibility leads to particle polymorphism. Linker flexibility may influence the dimeric-spike arrangement. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6999.map.gz emd_6999.map.gz | 21.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6999-v30.xml emd-6999-v30.xml emd-6999.xml emd-6999.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6999_fsc.xml emd_6999_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_6999.png emd_6999.png | 88.7 KB | ||
| Filedesc metadata |  emd-6999.cif.gz emd-6999.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6999 http://ftp.pdbj.org/pub/emdb/structures/EMD-6999 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6999 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6999 | HTTPS FTP |
-Related structure data
| Related structure data |  6ab5MC  9576C  5ykuC  5ykvC  5ykxC  5ykzC  5yl0C  5yl1C  6ab6C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6999.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6999.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.12 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Penaeus vannamei nodavirus
| Entire | Name:  Penaeus vannamei nodavirus Penaeus vannamei nodavirus |
|---|---|
| Components |
|
-Supramolecule #1: Penaeus vannamei nodavirus
| Supramolecule | Name: Penaeus vannamei nodavirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 430911 / Sci species name: Penaeus vannamei nodavirus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Litopenaeus vannamei (Pacific white shrimp) Litopenaeus vannamei (Pacific white shrimp) |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Penaeus vannamei nodavirus Penaeus vannamei nodavirus |
| Molecular weight | Theoretical: 40.262168 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKRKPNSNQN NNNNRGNGNG LRRVRGGRVS RRRVVINQSN QSMPVTSNGA PLQALTSYSR PNVNKISRLG PDSDFLTSVV AKASTSIVT PADRILVKQP LSASSFPGTR ITGLSSYWER YKWLSAVARY VPAVPNTVAC QFVMYIDTDP LDDPSNISDD N QIVRQAVS ...String: MKRKPNSNQN NNNNRGNGNG LRRVRGGRVS RRRVVINQSN QSMPVTSNGA PLQALTSYSR PNVNKISRLG PDSDFLTSVV AKASTSIVT PADRILVKQP LSASSFPGTR ITGLSSYWER YKWLSAVARY VPAVPNTVAC QFVMYIDTDP LDDPSNISDD N QIVRQAVS QAGSNQFNFN TSKTVPLIVR ADNQYYYTGV DKQNLRFSLQ GILYIIQVTD LINFNGELIT QDLTCGSLFL DW LVNFSIP QINPTSLTDV RVDKAVNFIK PEVSGVAEIQ TVTGLSPSTS YLLTPAFLEQ NFQSEAGIYI LSATPVEGEG TIS INMDPT VTTVSGFIKV KTDTFGTFDL SVVLTTASKK QTTGFNIIAA TS UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: MOLYBDENUM / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 2806 / Average exposure time: 2.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.75 µm / Nominal defocus min: 1.25 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)