[English] 日本語
 Yorodumi
Yorodumi- EMDB-6993: The cryo-EM structure of filamentous bacteriophage IKe applied wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6993 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of filamentous bacteriophage IKe applied with C5 and helical parameters. | ||||||||||||
 Map data Map data | The cryo-EM structure of filamentous bacteriophage IKe applied with helical parameters and 5-fold symmetry. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | filamentous bacteriophage / major coat protein / VIRAL PROTEIN | ||||||||||||
| Function / homology | Phage major coat protein, Gp8 / Bacteriophage M13, G8P, capsid domain superfamily / Capsid protein G8P / helical viral capsid / host cell membrane / Capsid protein G8P Function and homology information Function and homology information | ||||||||||||
| Biological species |  Filamentous phage (virus) Filamentous phage (virus) | ||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Xu JW / Dayan N | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Cryo-electron microscopy structure of the filamentous bacteriophage IKe. Authors: Jingwei Xu / Nir Dayan / Amir Goldbourt / Ye Xiang /   Abstract: The filamentous bacteriophage IKe infects cells bearing IncN pili. We report the cryo-electron microscopy structure of the micrometer-long IKe viral particle at a resolution of 3.4 Å. The major ...The filamentous bacteriophage IKe infects cells bearing IncN pili. We report the cryo-electron microscopy structure of the micrometer-long IKe viral particle at a resolution of 3.4 Å. The major coat protein [protein 8 (p8)] consists of 47 residues that fold into a ∼68-Å-long helix. An atomic model of the coat protein was built. Five p8 helices in a horizontal layer form a pentamer, and symmetrically neighboring p8 layers form a right-handed helical cylinder having a rise per pentamer of 16.77 Å and a twist of 38.52°. The inner surface of the capsid cylinder is positively charged and has direct interactions with the encapsulated circular single-stranded DNA genome, which has an electron density consistent with an unusual left-handed helix structure. Similar to capsid structures of other filamentous viruses, strong capsid packing in the IKe particle is maintained by hydrophobic residues. Despite having a different length and large sequence differences from other filamentous phages, π-π interactions were found between Tyr9 of one p8 and Trp29 of a neighboring p8 in IKe that are similar to interactions observed in phage M13, suggesting that, despite sequence divergence, overall structural features are maintained. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6993.map.gz emd_6993.map.gz | 3.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6993-v30.xml emd-6993-v30.xml emd-6993.xml emd-6993.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6993.png emd_6993.png | 34.8 KB | ||
| Filedesc metadata |  emd-6993.cif.gz emd-6993.cif.gz | 5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6993 http://ftp.pdbj.org/pub/emdb/structures/EMD-6993 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6993 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6993 | HTTPS FTP |
-Related structure data
| Related structure data |  6a7fMC  6994C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6993.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6993.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The cryo-EM structure of filamentous bacteriophage IKe applied with helical parameters and 5-fold symmetry. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
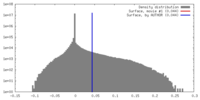
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Filamentous phage
| Entire | Name:  Filamentous phage (virus) Filamentous phage (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Filamentous phage
| Supramolecule | Name: Filamentous phage / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 12420 / Sci species name: Filamentous phage / Sci species strain: IKe / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Diameter: 64.0 Å |
-Macromolecule #1: major coat protein p8
| Macromolecule | Name: major coat protein p8 / type: protein_or_peptide / ID: 1 / Number of copies: 30 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Filamentous phage (virus) / Strain: IKe Filamentous phage (virus) / Strain: IKe |
| Molecular weight | Theoretical: 5.700578 KDa |
| Sequence | String: AEPNAATNYA TEAMDSLKTQ AIDLISQTWP VVTTVVVAGL VIRLFKKFSS KAV UniProtKB: Capsid protein G8P |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50mM Tris at pH 7.5, 10mM MgCl2, 100mM NaCl |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 16.77 Å Applied symmetry - Helical parameters - Δ&Phi: 38.52 ° Applied symmetry - Helical parameters - Axial symmetry: C5 (5 fold cyclic) Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 2.0) / Number images used: 112808 |
|---|---|
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: RELION (ver. 2) |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6a7f: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)