+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6876 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Methanoccus maripaludis archaellum | |||||||||
 Map data Map data | Methanococcus maripaludis archaellum cryo-EM reconstruction | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Archaellum / archea / flagellum / metal binding / motility / helical / PROTEIN FIBRIL | |||||||||
| Function / homology | archaeal-type flagellum / Flagellin, archaea / Archaebacterial flagellin / Flagellin/pilin, N-terminal / archaeal or bacterial-type flagellum-dependent cell motility / structural molecule activity / membrane / Flagellin Function and homology information Function and homology information | |||||||||
| Biological species |  Methanococcus maripaludis S2 (archaea) / Methanococcus maripaludis S2 (archaea) /  Methanococcus maripaludis (strain S2 / LL) (archaea) Methanococcus maripaludis (strain S2 / LL) (archaea) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Meshcheryakov VA / Shibata S / Schreiber MT / Villar-Briones A / Jarrell KF / Aizawa S / Wolf M / Kurumizaka H | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2019 Journal: EMBO Rep / Year: 2019Title: High-resolution archaellum structure reveals a conserved metal-binding site. Authors: Vladimir A Meshcheryakov / Satoshi Shibata / Makoto Tokoro Schreiber / Alejandro Villar-Briones / Kenneth F Jarrell / Shin-Ichi Aizawa / Matthias Wolf /   Abstract: Many archaea swim by means of archaella. While the archaellum is similar in function to its bacterial counterpart, its structure, composition, and evolution are fundamentally different. Archaella are ...Many archaea swim by means of archaella. While the archaellum is similar in function to its bacterial counterpart, its structure, composition, and evolution are fundamentally different. Archaella are related to archaeal and bacterial type IV pili. Despite recent advances, our understanding of molecular processes governing archaellum assembly and stability is still incomplete. Here, we determine the structures of archaella by X-ray crystallography and cryo-EM The crystal structure of FlaB1 is the first and only crystal structure of any archaellin to date at a resolution of 1.5 Å, which is put into biological context by a cryo-EM reconstruction from archaella at 4 Å resolution created with helical single-particle analysis. Our results indicate that the archaellum is predominantly composed of FlaB1. We identify N-linked glycosylation by cryo-EM and mass spectrometry. The crystal structure reveals a highly conserved metal-binding site, which is validated by mass spectrometry and electron energy-loss spectroscopy. We show that the metal-binding site, which appears to be a widespread property of archaellin, is required for filament integrity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6876.map.gz emd_6876.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6876-v30.xml emd-6876-v30.xml emd-6876.xml emd-6876.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6876.png emd_6876.png | 119.9 KB | ||
| Filedesc metadata |  emd-6876.cif.gz emd-6876.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6876 http://ftp.pdbj.org/pub/emdb/structures/EMD-6876 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6876 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6876 | HTTPS FTP |
-Validation report
| Summary document |  emd_6876_validation.pdf.gz emd_6876_validation.pdf.gz | 517.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6876_full_validation.pdf.gz emd_6876_full_validation.pdf.gz | 516.8 KB | Display | |
| Data in XML |  emd_6876_validation.xml.gz emd_6876_validation.xml.gz | 5.5 KB | Display | |
| Data in CIF |  emd_6876_validation.cif.gz emd_6876_validation.cif.gz | 6.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6876 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6876 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6876 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6876 | HTTPS FTP |
-Related structure data
| Related structure data |  5z1lMC  5ya6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6876.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6876.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Methanococcus maripaludis archaellum cryo-EM reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.41 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
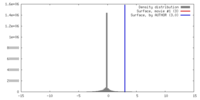
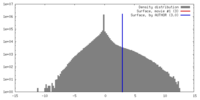
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : M.maripaludis archaellin FlaB1 filament
| Entire | Name: M.maripaludis archaellin FlaB1 filament |
|---|---|
| Components |
|
-Supramolecule #1: M.maripaludis archaellin FlaB1 filament
| Supramolecule | Name: M.maripaludis archaellin FlaB1 filament / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Methanococcus maripaludis S2 (archaea) Methanococcus maripaludis S2 (archaea) |
-Macromolecule #1: Flagellin
| Macromolecule | Name: Flagellin / type: protein_or_peptide / ID: 1 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Methanococcus maripaludis (strain S2 / LL) (archaea) Methanococcus maripaludis (strain S2 / LL) (archaea)Strain: S2 / LL |
| Molecular weight | Theoretical: 21.639359 KDa |
| Sequence | String: MKITEFMKSK KGASGIGTLI VFIAMVLVAA VAASVLINTS GFLQQKASTT GKESTEQVAS GLLINGITGS VGTSDVKLLA IYLAPNAGS SAIDLAQTKV MLDYNGKSVV LGYGGNQDMS SGNSSVFSND TGATATTFQV NILQDYDDSA VDNAVINKGD A VALIVDVN ...String: MKITEFMKSK KGASGIGTLI VFIAMVLVAA VAASVLINTS GFLQQKASTT GKESTEQVAS GLLINGITGS VGTSDVKLLA IYLAPNAGS SAIDLAQTKV MLDYNGKSVV LGYGGNQDMS SGNSSVFSND TGATATTFQV NILQDYDDSA VDNAVINKGD A VALIVDVN ASFAGEIPER TAISGKVQPE FGAPGVISFT TPASYTTTLV ELQ UniProtKB: Flagellin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | .8 mg/mL |
|---|---|
| Buffer | pH: 7 / Component - Formula: H2O / Component - Name: Water |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 100 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 9.33 kPa / Details: Gatan Solarus |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV / Details: 3 second blot, 3.5uL. |
| Details | sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 77.0 K / Max: 100.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.1 mrad |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Details | nanoprobe, parallel beam illumination |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7676 pixel / Digitization - Dimensions - Height: 7420 pixel / Digitization - Frames/image: 1-48 / Number grids imaged: 1 / Number real images: 2000 / Average exposure time: 12.0 sec. / Average electron dose: 96.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: -2.5 µm / Calibrated defocus min: -1.5 µm / Calibrated magnification: 47619 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.5 µm / Nominal defocus min: -1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-5z1l: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)