登録情報 データベース : EMDB / ID : EMD-6792タイトル Cryo-EM structure of p300 Cryo-EM structure of p300. Transcriptional coactivator p300 is a large, multidomain protein that possess (HAT) histone acetyltransferase activity. 細胞器官・細胞要素 : p300 proteinタンパク質・ペプチド : Histone acetyltransferase p300機能・相同性 分子機能 ドメイン・相同性 構成要素
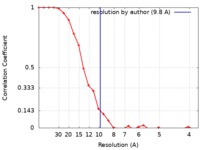
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト)手法 / / 解像度 : 9.8 Å Ghosh R / Roy S / Sengupta J ジャーナル : Biochemistry / 年 : 2019タイトル : Tumor Suppressor p53-Mediated Structural Reorganization of the Transcriptional Coactivator p300.著者 : Raka Ghosh / Stephanie Kaypee / Manidip Shasmal / Tapas K Kundu / Siddhartha Roy / Jayati Sengupta / 要旨 : Transcriptional coactivator p300, a critical player in eukaryotic gene regulation, primarily functions as a histone acetyltransferase (HAT). It is also an important player in acetylation of a number ... Transcriptional coactivator p300, a critical player in eukaryotic gene regulation, primarily functions as a histone acetyltransferase (HAT). It is also an important player in acetylation of a number of nonhistone proteins, p53 being the most prominent one. Recruitment of p300 to p53 is pivotal in the regulation of p53-dependent genes. Emerging evidence suggests that p300 adopts an active conformation upon binding to the tetrameric p53, resulting in its enhanced acetylation activity. As a modular protein, p300 consists of multiple well-defined domains, where the structured domains are interlinked with unstructured linker regions. A crystal structure of the central domain of p300 encompassing Bromo, RING, PHD, and HAT domains demonstrates a compact module, where the HAT active site stays occluded by the RING domain. However, although p300 has a significant role in mediating the transcriptional activity of p53, only a few structural details on the complex of these two full-length proteins are available. Here, we present a cryo-electron microscopy (cryo-EM) study on the p300-p53 complex. The three-dimensional cryo-EM density map of the p300-p53 complex, when compared to the cryo-EM map of free p300, revealed that substantial change in the relative arrangement of Bromo and HAT domains occurs upon complex formation, which is likely required for exposing HAT active site and subsequent acetyltransferase activity. Our observation correlates well with previous studies showing that the presence of Bromodomain is obligatory for effective acetyltransferase activity of HAT. Thus, our result sheds new light on the mechanism whereby p300, following binding with p53, gets activated. 履歴 登録 2017年7月13日 - ヘッダ(付随情報) 公開 2019年1月23日 - マップ公開 2019年1月23日 - 更新 2020年1月29日 - 現状 2020年1月29日 処理サイト : PDBj / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) データ登録者
データ登録者 引用
引用 ジャーナル: Biochemistry / 年: 2019
ジャーナル: Biochemistry / 年: 2019
 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_6792.map.gz
emd_6792.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-6792-v30.xml
emd-6792-v30.xml emd-6792.xml
emd-6792.xml EMDBヘッダ
EMDBヘッダ emd_6792_fsc.xml
emd_6792_fsc.xml FSCデータファイル
FSCデータファイル emd_6792.png
emd_6792.png http://ftp.pdbj.org/pub/emdb/structures/EMD-6792
http://ftp.pdbj.org/pub/emdb/structures/EMD-6792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6792
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6792 emd_6792_validation.pdf.gz
emd_6792_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_6792_full_validation.pdf.gz
emd_6792_full_validation.pdf.gz emd_6792_validation.xml.gz
emd_6792_validation.xml.gz emd_6792_validation.cif.gz
emd_6792_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6792
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6792 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6792
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6792 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_6792.map.gz / 形式: CCP4 / 大きさ: 1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_6792.map.gz / 形式: CCP4 / 大きさ: 1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素 Homo sapiens (ヒト)
Homo sapiens (ヒト)
 Homo sapiens (ヒト)
Homo sapiens (ヒト)
 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 ムービー
ムービー コントローラー
コントローラー







































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)