[English] 日本語
 Yorodumi
Yorodumi- EMDB-6731: Anti-CRISPR proteins AcrF1/2 bound to Csy surveillance complex wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6731 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Anti-CRISPR proteins AcrF1/2 bound to Csy surveillance complex with a 20nt spacer crRNA backbone region | |||||||||
 Map data Map data | anti-CRISPR proteins AcrF1/2 bound to Csy surveillance complex with 20nt spacer crRNA backbone region | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | anti-CRISPR proteins / Csy complex / Type I-F CRISPR/Cas system / IMMUNE SYSTEM-RNA complex | |||||||||
| Function / homology | : / Anti-CRISPR protein Acr30-35/AcrF1 / CRISPR-associated protein Csy3 / CRISPR-associated protein (Cas_Csy3) / defense response to virus / Uncharacterized protein / CRISPR-associated protein Csy3 Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) / Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) /  Pseudomonas phage JBD30 (virus) / Pseudomonas phage JBD30 (virus) /  | |||||||||
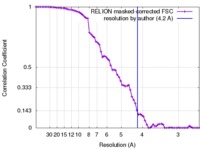
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Peng R / Shi Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2017 Journal: Cell Res / Year: 2017Title: Alternate binding modes of anti-CRISPR viral suppressors AcrF1/2 to Csy surveillance complex revealed by cryo-EM structures. Authors: Ruchao Peng / Ying Xu / Tengfei Zhu / Ningning Li / Jianxun Qi / Yan Chai / Min Wu / Xinzheng Zhang / Yi Shi / Peiyi Wang / Jiawei Wang / Ning Gao / George Fu Gao /    Abstract: Bacteriophages encode anti-CRISPR suppressors to counteract the CRISPR/Cas immunity of their bacterial hosts, thus facilitating their survival and replication. Previous studies have shown that two ...Bacteriophages encode anti-CRISPR suppressors to counteract the CRISPR/Cas immunity of their bacterial hosts, thus facilitating their survival and replication. Previous studies have shown that two phage-encoded anti-CRISPR proteins, AcrF1 and AcrF2, suppress the type I-F CRISPR/Cas system of Pseudomonas aeruginosa by preventing target DNA recognition by the Csy surveillance complex, but the precise underlying mechanism was unknown. Here we present the structure of AcrF1/2 bound to the Csy complex determined by cryo-EM single-particle reconstruction. By structural analysis, we found that AcrF1 inhibits target DNA recognition of the Csy complex by interfering with base pairing between the DNA target strand and crRNA spacer. In addition, multiple copies of AcrF1 bind to the Csy complex with different modes when working individually or cooperating with AcrF2, which might exclude target DNA binding through different mechanisms. Together with previous reports, we provide a comprehensive working scenario for the two anti-CRISPR suppressors, AcrF1 and AcrF2, which silence CRISPR/Cas immunity by targeting the Csy surveillance complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6731.map.gz emd_6731.map.gz | 2.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6731-v30.xml emd-6731-v30.xml emd-6731.xml emd-6731.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6731_fsc.xml emd_6731_fsc.xml | 7 KB | Display |  FSC data file FSC data file |
| Images |  emd_6731.png emd_6731.png | 152 KB | ||
| Filedesc metadata |  emd-6731.cif.gz emd-6731.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6731 http://ftp.pdbj.org/pub/emdb/structures/EMD-6731 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6731 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6731 | HTTPS FTP |
-Related structure data
| Related structure data |  5xlpMC  6728C  6729C  6730C  5xloC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6731.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6731.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | anti-CRISPR proteins AcrF1/2 bound to Csy surveillance complex with 20nt spacer crRNA backbone region | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
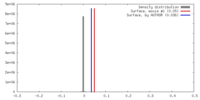
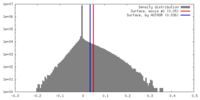
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : anti-CRISPR proteins AcrF1/2 bound to Csy surveillance complex wi...
| Entire | Name: anti-CRISPR proteins AcrF1/2 bound to Csy surveillance complex with a 20nt spacer sequence backbone region |
|---|---|
| Components |
|
-Supramolecule #1: anti-CRISPR proteins AcrF1/2 bound to Csy surveillance complex wi...
| Supramolecule | Name: anti-CRISPR proteins AcrF1/2 bound to Csy surveillance complex with a 20nt spacer sequence backbone region type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)Strain: UCBPP-PA14 |
| Molecular weight | Theoretical: 300 KDa |
-Supramolecule #2: man CRISPR-associated protein Csy3
| Supramolecule | Name: man CRISPR-associated protein Csy3 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|
-Supramolecule #3: RNA
| Supramolecule | Name: RNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Supramolecule #4: AcrF1
| Supramolecule | Name: AcrF1 / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|
-Macromolecule #1: CRISPR-associated protein Csy3
| Macromolecule | Name: CRISPR-associated protein Csy3 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)Strain: UCBPP-PA14 |
| Molecular weight | Theoretical: 37.579273 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKPILSTAS VLAFERKLDP SDALMSAGAW AQRDASQEWP AVTVREKSVR GTISNRLKTK DRDPAKLDAS IQSPNLQTVD VANLPSDAD TLKVRFTLRV LGGAGTPSAC NDAAYRDKLL QTVATYVNDQ GFAELARRYA HNLANARFLW RNRVGAEAVE V RINHIRQG ...String: MSKPILSTAS VLAFERKLDP SDALMSAGAW AQRDASQEWP AVTVREKSVR GTISNRLKTK DRDPAKLDAS IQSPNLQTVD VANLPSDAD TLKVRFTLRV LGGAGTPSAC NDAAYRDKLL QTVATYVNDQ GFAELARRYA HNLANARFLW RNRVGAEAVE V RINHIRQG EVARAWRFDA LAIGLRDFKA DAELDALAEL IASGLSGSGH VLLEVVAFAR IGDGQEVFPS QELILDKGDK KG QKSKTLY SVRDAAAIHS QKIGNALRTI DTWYPDEDGL GPIAVEPYGS VTSQGKAYRQ PKQKLDFYTL LDNWVLRDEA PAV EQQHYV IANLIRGGVF GEAEEK UniProtKB: CRISPR-associated protein Csy3 |
-Macromolecule #3: Uncharacterized protein AcrF1
| Macromolecule | Name: Uncharacterized protein AcrF1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage JBD30 (virus) Pseudomonas phage JBD30 (virus) |
| Molecular weight | Theoretical: 8.824931 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKFIKYLSTA HLNYMNIAVY ENGSKIKARV ENVVNGKSVG ARDFDSTEQL ESWFYGLPGS GLGRIENAMN EISRRENP UniProtKB: Uncharacterized protein |
-Macromolecule #2: crRNA with 20nt spacer sequence
| Macromolecule | Name: crRNA with 20nt spacer sequence / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.456173 KDa |
| Sequence | String: CUAAGAAAUU CACGGCGGGC UUGAUGUCGU UCACUGCCGU GUAGGCAG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 0.35 sec. / Average electron dose: 1.55 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-5xlp: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)