+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6690 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Apaf-1-Caspase-9 holoenzyme | ||||||||||||
 Map data Map data | Apaf-1-Caspase-9 holoenzyme platform | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | apoptosis holoenzyme / APOPTOSIS | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationcaspase-9 / response to G1 DNA damage checkpoint signaling / caspase complex / regulation of apoptotic DNA fragmentation / cytochrome c-heme linkage / Formation of apoptosome / apoptosome / cytochrome complex / leukocyte apoptotic process / glial cell apoptotic process ...caspase-9 / response to G1 DNA damage checkpoint signaling / caspase complex / regulation of apoptotic DNA fragmentation / cytochrome c-heme linkage / Formation of apoptosome / apoptosome / cytochrome complex / leukocyte apoptotic process / glial cell apoptotic process / response to cobalt ion / cysteine-type endopeptidase activator activity / Caspase activation via Dependence Receptors in the absence of ligand / Activation of caspases through apoptosome-mediated cleavage / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes / Regulation of the apoptosome activity / fibroblast apoptotic process / AKT phosphorylates targets in the cytosol / epithelial cell apoptotic process / mitochondrial electron transport, cytochrome c to oxygen / platelet formation / response to anesthetic / cysteine-type endopeptidase activator activity involved in apoptotic process / mitochondrial electron transport, ubiquinol to cytochrome c / TP53 Regulates Transcription of Caspase Activators and Caspases / Constitutive Signaling by AKT1 E17K in Cancer / Transcriptional Regulation by E2F6 / forebrain development / intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress / positive regulation of execution phase of apoptosis / cellular response to dexamethasone stimulus / cellular response to transforming growth factor beta stimulus / response to UV / signal transduction in response to DNA damage / heat shock protein binding / cardiac muscle cell apoptotic process / response to nutrient / intrinsic apoptotic signaling pathway / response to ischemia / positive regulation of apoptotic signaling pathway / protein maturation / neural tube closure / kidney development / apoptotic signaling pathway / enzyme activator activity / NOD1/2 Signaling Pathway / protein processing / ADP binding / mitochondrial intermembrane space / SH3 domain binding / intrinsic apoptotic signaling pathway in response to DNA damage / cellular response to UV / response to estradiol / peptidase activity / nervous system development / positive regulation of neuron apoptotic process / neuron apoptotic process / secretory granule lumen / regulation of apoptotic process / response to lipopolysaccharide / ficolin-1-rich granule lumen / response to hypoxia / cell differentiation / electron transfer activity / positive regulation of apoptotic process / cysteine-type endopeptidase activity / nucleotide binding / apoptotic process / heme binding / DNA damage response / Neutrophil degranulation / lipid binding / protein kinase binding / protein-containing complex / mitochondrion / proteolysis / extracellular exosome / extracellular region / ATP binding / metal ion binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | ||||||||||||
 Authors Authors | Li Y / Zhou M | ||||||||||||
| Funding support |  China, China,  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme. Authors: Yini Li / Mengying Zhou / Qi Hu / Xiao-Chen Bai / Weiyun Huang / Sjors H W Scheres / Yigong Shi /   Abstract: Mammalian intrinsic apoptosis requires activation of the initiator caspase-9, which then cleaves and activates the effector caspases to execute cell killing. The heptameric Apaf-1 apoptosome is ...Mammalian intrinsic apoptosis requires activation of the initiator caspase-9, which then cleaves and activates the effector caspases to execute cell killing. The heptameric Apaf-1 apoptosome is indispensable for caspase-9 activation by together forming a holoenzyme. The molecular mechanism of caspase-9 activation remains largely enigmatic. Here, we report the cryoelectron microscopy (cryo-EM) structure of an apoptotic holoenzyme and structure-guided biochemical analyses. The caspase recruitment domains (CARDs) of Apaf-1 and caspase-9 assemble in two different ways: a 4:4 complex docks onto the central hub of the apoptosome, and a 2:1 complex binds the periphery of the central hub. The interface between the CARD complex and the central hub is required for caspase-9 activation within the holoenzyme. Unexpectedly, the CARD of free caspase-9 strongly inhibits its proteolytic activity. These structural and biochemical findings demonstrate that the apoptosome activates caspase-9 at least in part through sequestration of the inhibitory CARD domain. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6690.map.gz emd_6690.map.gz | 117 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6690-v30.xml emd-6690-v30.xml emd-6690.xml emd-6690.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6690.png emd_6690.png | 94.4 KB | ||
| Filedesc metadata |  emd-6690.cif.gz emd-6690.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6690 http://ftp.pdbj.org/pub/emdb/structures/EMD-6690 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6690 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6690 | HTTPS FTP |
-Related structure data
| Related structure data |  5wveMC  6691C  6692C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6690.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6690.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Apaf-1-Caspase-9 holoenzyme platform | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
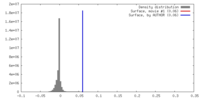
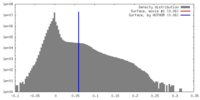
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Apaf-1 apoptosome
+Supramolecule #1: Apaf-1 apoptosome
+Supramolecule #2: Apaf-1
+Supramolecule #3: cytochrome c
+Supramolecule #4: Apaf-1 CARD
+Supramolecule #5: caspase-9 CARD
+Macromolecule #1: Apoptotic protease-activating factor 1
+Macromolecule #2: Cytochrome c
+Macromolecule #3: Apoptotic protease-activating factor 1
+Macromolecule #4: Caspase
+Macromolecule #5: 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE
+Macromolecule #6: MAGNESIUM ION
+Macromolecule #7: PROTOPORPHYRIN IX CONTAINING FE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 240130 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)