[English] 日本語
 Yorodumi
Yorodumi- EMDB-6588: Negative stain EM of Ebola virus GP bound to human survivor antibodies -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6588 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
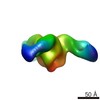
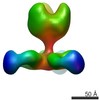
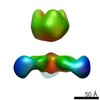
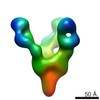
| Title | Negative stain EM of Ebola virus GP bound to human survivor antibodies | |||||||||
 Map data Map data | Reconstruction of human survivor antibody Fabs bound to EBOV GPdTM | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ebola virus / antibodies / negative stain EM | |||||||||
| Biological species |  Ebola virus sp. / Ebola virus sp. /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Borndoldt ZA / Turner HL / Murin CD / Li W / Sok D / Sounders C / Piper AE / Fusco ML / Pommert K / Klempner M ...Borndoldt ZA / Turner HL / Murin CD / Li W / Sok D / Sounders C / Piper AE / Fusco ML / Pommert K / Klempner M / Reimann KA / Golf A / Wittrup DK / Gerngross TU / Krauland E / Glass PJ / Burton DR / Saphire EO / Ward AB / Walker LM | |||||||||
 Citation Citation |  Journal: Science / Year: 2016 Journal: Science / Year: 2016Title: Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Authors: Zachary A Bornholdt / Hannah L Turner / Charles D Murin / Wen Li / Devin Sok / Colby A Souders / Ashley E Piper / Arthur Goff / Joshua D Shamblin / Suzanne E Wollen / Thomas R Sprague / ...Authors: Zachary A Bornholdt / Hannah L Turner / Charles D Murin / Wen Li / Devin Sok / Colby A Souders / Ashley E Piper / Arthur Goff / Joshua D Shamblin / Suzanne E Wollen / Thomas R Sprague / Marnie L Fusco / Kathleen B J Pommert / Lisa A Cavacini / Heidi L Smith / Mark Klempner / Keith A Reimann / Eric Krauland / Tillman U Gerngross / Karl D Wittrup / Erica Ollmann Saphire / Dennis R Burton / Pamela J Glass / Andrew B Ward / Laura M Walker /  Abstract: Antibodies targeting the Ebola virus surface glycoprotein (EBOV GP) are implicated in protection against lethal disease, but the characteristics of the human antibody response to EBOV GP remain ...Antibodies targeting the Ebola virus surface glycoprotein (EBOV GP) are implicated in protection against lethal disease, but the characteristics of the human antibody response to EBOV GP remain poorly understood. We isolated and characterized 349 GP-specific monoclonal antibodies (mAbs) from the peripheral B cells of a convalescent donor who survived the 2014 EBOV Zaire outbreak. Remarkably, 77% of the mAbs neutralize live EBOV, and several mAbs exhibit unprecedented potency. Structures of selected mAbs in complex with GP reveal a site of vulnerability located in the GP stalk region proximal to the viral membrane. Neutralizing antibodies targeting this site show potent therapeutic efficacy against lethal EBOV challenge in mice. The results provide a framework for the design of new EBOV vaccine candidates and immunotherapies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6588.map.gz emd_6588.map.gz | 893.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6588-v30.xml emd-6588-v30.xml emd-6588.xml emd-6588.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6588.jpg emd_6588.jpg | 18.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6588 http://ftp.pdbj.org/pub/emdb/structures/EMD-6588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6588 | HTTPS FTP |
-Validation report
| Summary document |  emd_6588_validation.pdf.gz emd_6588_validation.pdf.gz | 78.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6588_full_validation.pdf.gz emd_6588_full_validation.pdf.gz | 77.9 KB | Display | |
| Data in XML |  emd_6588_validation.xml.gz emd_6588_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6588 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6588 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6588 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6588 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6588.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6588.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of human survivor antibody Fabs bound to EBOV GPdTM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Reconstruction of Ebola virus GP ectodomain containing mucin-like...
| Entire | Name: Reconstruction of Ebola virus GP ectodomain containing mucin-like domains in complex with human survivor antibody Fab ADI-15758 |
|---|---|
| Components |
|
-Supramolecule #1000: Reconstruction of Ebola virus GP ectodomain containing mucin-like...
| Supramolecule | Name: Reconstruction of Ebola virus GP ectodomain containing mucin-like domains in complex with human survivor antibody Fab ADI-15758 type: sample / ID: 1000 / Oligomeric state: One GP trimer bound to three Fab monomers / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: Ebola virus glycoprotein
| Macromolecule | Name: Ebola virus glycoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: EBOV GPdTM / Number of copies: 1 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Ebola virus sp. / synonym: Ebola virus Ebola virus sp. / synonym: Ebola virus |
| Molecular weight | Theoretical: 210 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #2: Immunoglobulin 1 fragment
| Macromolecule | Name: Immunoglobulin 1 fragment / type: protein_or_peptide / ID: 2 / Name.synonym: IgG1 Fab / Number of copies: 3 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 50 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 150 mM NaCl, 20 mM Tris |
| Staining | Type: NEGATIVE / Details: 2% uranyl formate |
| Grid | Details: Cu mesh grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Date | Sep 10, 2015 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Number real images: 98 / Average electron dose: 25 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Particles were picked using DoG Picker. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: OTHER / Software - Name: EMAN2, XMipp, IMAGIC / Number images used: 17771 |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera