[English] 日本語
 Yorodumi
Yorodumi- EMDB-6585: The structure of elongation factor 4 (EF4/LepA) in GTP form bound... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6585 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of elongation factor 4 (EF4/LepA) in GTP form bound to the ribosome | |||||||||
 Map data Map data | Reconstruction of EF4 bound to the 70S ribosome | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | LepA / EF4 / ribosome / 70S | |||||||||
| Function / homology |  Function and homology information Function and homology information: / translation elongation factor activity / positive regulation of translation / regulation of translation / large ribosomal subunit / transferase activity / ribosome binding / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding ...: / translation elongation factor activity / positive regulation of translation / regulation of translation / large ribosomal subunit / transferase activity / ribosome binding / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / GTPase activity / GTP binding / zinc ion binding / metal ion binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.9 Å | |||||||||
 Authors Authors | Kumar V / Ahmed T / Ero R / Jian G-K / Yin Z / Gao Y / Bhushan S | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2016 Journal: J Biol Chem / Year: 2016Title: Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome. Authors: Veerendra Kumar / Rya Ero / Tofayel Ahmed / Kwok Jian Goh / Yin Zhan / Shashi Bhushan / Yong-Gui Gao /  Abstract: Elongation factor 4 (EF4) is a member of the family of ribosome-dependent translational GTPase factors, along with elongation factor G and BPI-inducible protein A. Although EF4 is highly conserved in ...Elongation factor 4 (EF4) is a member of the family of ribosome-dependent translational GTPase factors, along with elongation factor G and BPI-inducible protein A. Although EF4 is highly conserved in bacterial, mitochondrial, and chloroplast genomes, its exact biological function remains controversial. Here we present the cryo-EM reconstitution of the GTP form of EF4 bound to the ribosome with P and E site tRNAs at 3.8-Å resolution. Interestingly, our structure reveals an unrotated ribosome rather than a clockwise-rotated ribosome, as observed in the presence of EF4-GDP and P site tRNA. In addition, we also observed a counterclockwise-rotated form of the above complex at 5.7-Å resolution. Taken together, our results shed light on the interactions formed between EF4, the ribosome, and the P site tRNA and illuminate the GTPase activation mechanism at previously unresolved detail. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6585.map.gz emd_6585.map.gz | 12.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6585-v30.xml emd-6585-v30.xml emd-6585.xml emd-6585.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6585.gif 400_6585.gif 80_6585.gif 80_6585.gif | 71.9 KB 5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6585 http://ftp.pdbj.org/pub/emdb/structures/EMD-6585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6585 | HTTPS FTP |
-Related structure data
| Related structure data |  5imrMC  6584C  5imqC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6585.map.gz / Format: CCP4 / Size: 126.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6585.map.gz / Format: CCP4 / Size: 126.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of EF4 bound to the 70S ribosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.28 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
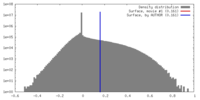
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The structure of elongation factor 4 (EF4/LepA) in GTP form bound...
| Entire | Name: The structure of elongation factor 4 (EF4/LepA) in GTP form bound to the ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: The structure of elongation factor 4 (EF4/LepA) in GTP form bound...
| Supramolecule | Name: The structure of elongation factor 4 (EF4/LepA) in GTP form bound to the ribosome type: sample / ID: 1000 / Oligomeric state: 1 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 2.5 MDa / Theoretical: 2.5 MDa / Method: Sedimentation |
-Supramolecule #1: Thermus thermophilus
| Supramolecule | Name: Thermus thermophilus / type: complex / ID: 1 / Name.synonym: 70S ribosome / Recombinant expression: No / Database: NCBI / Ribosome-details: ribosome-prokaryote: ALL |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) / Location in cell: Translation Thermus thermophilus (bacteria) / Location in cell: Translation |
| Molecular weight | Experimental: 2.5 MDa / Theoretical: 2.5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 5 mM HEPES, pH 7.5, 10 mM MgOAc, 50 mM KCl, 10 mM NH4Cl, 6 mM 2-mercaptoethanol |
|---|---|
| Grid | Details: 300 mesh copper grid with 2 nm carbon support, glow-discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV / Method: Blot for 3 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 73684 times magnification |
| Date | Jul 24, 2015 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 480 / Average electron dose: 20 e/Å2 Details: Every image is the average of seven frames recorded by the direct electron detector |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 109375 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 78000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Particles were selected using EMAN and processed using Relion |
|---|---|
| CTF correction | Details: Ctffind3 |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 5.9 Å / Resolution method: OTHER / Software - Name: Relion / Number images used: 18100 |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)