[English] 日本語
 Yorodumi
Yorodumi- EMDB-6542: Structure of Simian Immunodeficiency Virus Envelope Spikes bound ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6542 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5 | |||||||||
 Map data Map data | Reconstruction of closed state SIV envelope spike bound with CD4 and 36D5 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryoelectron tomography / image processing / electron microscopy / immunology / AIDS / HIV | |||||||||
| Function / homology |  Function and homology information Function and homology informationhelper T cell enhancement of adaptive immune response / interleukin-16 binding / interleukin-16 receptor activity / maintenance of protein location in cell / response to methamphetamine hydrochloride / cellular response to ionomycin / T cell selection / MHC class II protein binding / positive regulation of kinase activity / cellular response to granulocyte macrophage colony-stimulating factor stimulus ...helper T cell enhancement of adaptive immune response / interleukin-16 binding / interleukin-16 receptor activity / maintenance of protein location in cell / response to methamphetamine hydrochloride / cellular response to ionomycin / T cell selection / MHC class II protein binding / positive regulation of kinase activity / cellular response to granulocyte macrophage colony-stimulating factor stimulus / interleukin-15-mediated signaling pathway / positive regulation of monocyte differentiation / Alpha-defensins / Nef Mediated CD4 Down-regulation / regulation of T cell activation / response to vitamin D / extracellular matrix structural constituent / Other interleukin signaling / T cell receptor complex / enzyme-linked receptor protein signaling pathway / Translocation of ZAP-70 to Immunological synapse / Phosphorylation of CD3 and TCR zeta chains / positive regulation of protein kinase activity / regulation of calcium ion transport / macrophage differentiation / Generation of second messenger molecules / positive regulation of calcium ion transport into cytosol / immunoglobulin binding / T cell differentiation / Co-inhibition by PD-1 / Binding and entry of HIV virion / coreceptor activity / positive regulation of calcium-mediated signaling / positive regulation of T cell proliferation / positive regulation of interleukin-2 production / cell surface receptor protein tyrosine kinase signaling pathway / protein tyrosine kinase binding / Vpu mediated degradation of CD4 / clathrin-coated endocytic vesicle membrane / calcium-mediated signaling / positive regulation of protein phosphorylation / transmembrane signaling receptor activity / MHC class II protein complex binding / response to estradiol / Downstream TCR signaling / Cargo recognition for clathrin-mediated endocytosis / signaling receptor activity / Clathrin-mediated endocytosis / virus receptor activity / clathrin-dependent endocytosis of virus by host cell / response to ethanol / defense response to Gram-negative bacterium / adaptive immune response / positive regulation of viral entry into host cell / early endosome / cell surface receptor signaling pathway / positive regulation of canonical NF-kappaB signal transduction / positive regulation of ERK1 and ERK2 cascade / cell adhesion / positive regulation of MAPK cascade / immune response / membrane raft / endoplasmic reticulum lumen / external side of plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / lipid binding / protein kinase binding / endoplasmic reticulum membrane / positive regulation of DNA-templated transcription / virion attachment to host cell / virion membrane / enzyme binding / signal transduction / protein homodimerization activity / zinc ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Simian immunodeficiency virus Simian immunodeficiency virus | |||||||||
| Method | subtomogram averaging / cryo EM | |||||||||
 Authors Authors | Hu G / Liu J / Roux KH / Taylor KA | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2017 Journal: J Virol / Year: 2017Title: Structure of Simian Immunodeficiency Virus Envelope Spikes Bound with CD4 and Monoclonal Antibody 36D5. Authors: Guiqing Hu / Jun Liu / Kenneth H Roux / Kenneth A Taylor /  Abstract: The human immunodeficiency virus type 1 (HIV-1)/simian immunodeficiency virus (SIV) envelope spike (Env) mediates viral entry into host cells. The V3 loop of the gp120 component of the Env trimer ...The human immunodeficiency virus type 1 (HIV-1)/simian immunodeficiency virus (SIV) envelope spike (Env) mediates viral entry into host cells. The V3 loop of the gp120 component of the Env trimer contributes to the coreceptor binding site and is a target for neutralizing antibodies. We used cryo-electron tomography to visualize the binding of CD4 and the V3 loop monoclonal antibody (MAb) 36D5 to gp120 of the SIV Env trimer. Our results show that 36D5 binds gp120 at the base of the V3 loop and suggest that the antibody exerts its neutralization effect by blocking the coreceptor binding site. The antibody does this without altering the dynamics of the spike motion between closed and open states when CD4 is bound. The interaction between 36D5 and SIV gp120 is similar to the interaction between some broadly neutralizing anti-V3 loop antibodies and HIV-1 gp120. Two conformations of gp120 bound with CD4 are revealed, suggesting an intrinsic dynamic nature of the liganded Env trimer. CD4 binding substantially increases the binding of 36D5 to gp120 in the intact Env trimer, consistent with CD4-induced changes in the conformation of gp120 and the antibody binding site. Binding by MAb 36D5 does not substantially alter the proportions of the two CD4-bound conformations. The position of MAb 36D5 at the V3 base changes little between conformations, indicating that the V3 base serves as a pivot point during the transition between these two states. Glycoprotein spikes on the surfaces of SIV and HIV are the sole targets available to the immune system for antibody neutralization. Spikes evade the immune system by a combination of a thick layer of polysaccharide on the surface (the glycan shield) and movement between spike domains that masks the epitope conformation. Using SIV virions whose spikes were "decorated" with the primary cellular receptor (CD4) and an antibody (36D5) at part of the coreceptor binding site, we visualized multiple conformations trapped by the rapid freezing step, which were separated using statistical analysis. Our results show that the CD4-induced conformational dynamics of the spike enhances binding of the antibody. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6542.map.gz emd_6542.map.gz | 265.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6542-v30.xml emd-6542-v30.xml emd-6542.xml emd-6542.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6542.tif emd_6542.tif | 71.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6542 http://ftp.pdbj.org/pub/emdb/structures/EMD-6542 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6542 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6542 | HTTPS FTP |
-Related structure data
| Related structure data |  3jcbMC  6538C  6539C 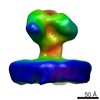 6540C 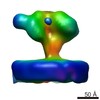 6541C  6543C  3jccC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6542.map.gz / Format: CCP4 / Size: 348.6 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6542.map.gz / Format: CCP4 / Size: 348.6 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of closed state SIV envelope spike bound with CD4 and 36D5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
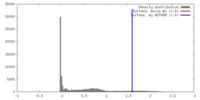
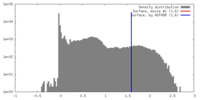
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SIV envelope bound to CD4 and monoclonal antibody 36D5 in closed state
| Entire | Name: SIV envelope bound to CD4 and monoclonal antibody 36D5 in closed state |
|---|---|
| Components |
|
-Supramolecule #1000: SIV envelope bound to CD4 and monoclonal antibody 36D5 in closed state
| Supramolecule | Name: SIV envelope bound to CD4 and monoclonal antibody 36D5 in closed state type: sample / ID: 1000 Oligomeric state: gp120 trimer bound to CD4 and one antibody 36D5 Number unique components: 3 |
|---|
-Supramolecule #1: Simian immunodeficiency virus
| Supramolecule | Name: Simian immunodeficiency virus / type: virus / ID: 1 / Name.synonym: SIV / NCBI-ID: 11723 / Sci species name: Simian immunodeficiency virus / Sci species strain: SIV239/251tail/Supt-CCR5 CL.30 / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No / Syn species name: SIV |
|---|---|
| Host (natural) | Organism:  Cercopithecidae (Old World monkeys) / synonym: VERTEBRATES Cercopithecidae (Old World monkeys) / synonym: VERTEBRATES |
-Macromolecule #1: T-cell surface glycoprotein CD4
| Macromolecule | Name: T-cell surface glycoprotein CD4 / type: protein_or_peptide / ID: 1 / Name.synonym: CD4 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human Homo sapiens (human) / synonym: human |
-Macromolecule #2: monoclonal antibody 36D5
| Macromolecule | Name: monoclonal antibody 36D5 / type: protein_or_peptide / ID: 2 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Date | Aug 16, 2008 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Average electron dose: 100 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 4.0 µm / Nominal magnification: 31000 |
| Sample stage | Specimen holder: cryo holder / Specimen holder model: OTHER / Tilt series - Axis1 - Min angle: -65 ° / Tilt series - Axis1 - Max angle: 65 ° |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Software - Name: I3, protomo / Number subtomograms used: 1181 |
|---|
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)