+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6490 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy structure of Apo-RAG (C2 symmetry) | |||||||||
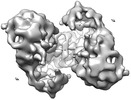
 Map data Map data | Reconstruction of Apo-RAG | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RAG1 / RAG2 / V(D)J recombination / Apo-RAG complex / Antigen receptor recombination / T and B cell development | |||||||||
| Function / homology |  Function and homology information Function and homology informationsomatic diversification of immune receptors via germline recombination within a single locus / protein binding / protein-DNA complex assembly / DNA recombinase complex / endodeoxyribonuclease complex / V(D)J recombination / phosphatidylinositol-3,4,5-trisphosphate binding / phosphatidylinositol-4,5-bisphosphate binding / B cell differentiation / RING-type E3 ubiquitin transferase ...somatic diversification of immune receptors via germline recombination within a single locus / protein binding / protein-DNA complex assembly / DNA recombinase complex / endodeoxyribonuclease complex / V(D)J recombination / phosphatidylinositol-3,4,5-trisphosphate binding / phosphatidylinositol-4,5-bisphosphate binding / B cell differentiation / RING-type E3 ubiquitin transferase / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / T cell differentiation in thymus / chromatin organization / endonuclease activity / histone binding / sequence-specific DNA binding / Hydrolases; Acting on ester bonds / adaptive immune response / chromatin binding / magnesium ion binding / protein homodimerization activity / DNA binding / zinc ion binding / metal ion binding / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Ru H / Chambers MG / Fu T / Tong AB / Liao M / Wu H | |||||||||
 Citation Citation |  Journal: Cell / Year: 2015 Journal: Cell / Year: 2015Title: Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Authors: Heng Ru / Melissa G Chambers / Tian-Min Fu / Alexander B Tong / Maofu Liao / Hao Wu /  Abstract: Diverse repertoires of antigen-receptor genes that result from combinatorial splicing of coding segments by V(D)J recombination are hallmarks of vertebrate immunity. The (RAG1-RAG2)2 recombinase (RAG) ...Diverse repertoires of antigen-receptor genes that result from combinatorial splicing of coding segments by V(D)J recombination are hallmarks of vertebrate immunity. The (RAG1-RAG2)2 recombinase (RAG) recognizes recombination signal sequences (RSSs) containing a heptamer, a spacer of 12 or 23 base pairs, and a nonamer (12-RSS or 23-RSS) and introduces precise breaks at RSS-coding segment junctions. RAG forms synaptic complexes only with one 12-RSS and one 23-RSS, a dogma known as the 12/23 rule that governs the recombination fidelity. We report cryo-electron microscopy structures of synaptic RAG complexes at up to 3.4 Å resolution, which reveal a closed conformation with base flipping and base-specific recognition of RSSs. Distortion at RSS-coding segment junctions and base flipping in coding segments uncover the two-metal-ion catalytic mechanism. Induced asymmetry involving tilting of the nonamer-binding domain dimer of RAG1 upon binding of HMGB1-bent 12-RSS or 23-RSS underlies the molecular mechanism for the 12/23 rule. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6490.map.gz emd_6490.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6490-v30.xml emd-6490-v30.xml emd-6490.xml emd-6490.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6490.png emd_6490.png | 335.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6490 http://ftp.pdbj.org/pub/emdb/structures/EMD-6490 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6490 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6490 | HTTPS FTP |
-Related structure data
| Related structure data |  6487C  6488C  6489C  6491C  3jbwC  3jbxC  3jbyC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6490.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6490.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of Apo-RAG | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
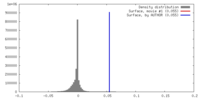
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Apo-RAG complex
| Entire | Name: Apo-RAG complex |
|---|---|
| Components |
|
-Supramolecule #1000: Apo-RAG complex
| Supramolecule | Name: Apo-RAG complex / type: sample / ID: 1000 / Details: The sample was monodisperse. / Oligomeric state: Dimer of RAG1-RAG2 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 290 KDa / Theoretical: 290 KDa / Method: Size exclusion chromatography |
-Macromolecule #1: Recombination Activating Gene 1 and 2
| Macromolecule | Name: Recombination Activating Gene 1 and 2 / type: protein_or_peptide / ID: 1 / Name.synonym: RAG1 and RAG2 / Number of copies: 2 / Oligomeric state: Dimer of RAG1-RAG2 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 290 KDa / Theoretical: 290 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: V(D)J recombination-activating protein 1 GO: DNA binding, endonuclease activity, ubiquitin-protein transferase activity, protein binding, zinc ion binding, DNA binding, chromatin binding, protein binding, phosphatidylinositol-4,5- ...GO: DNA binding, endonuclease activity, ubiquitin-protein transferase activity, protein binding, zinc ion binding, DNA binding, chromatin binding, protein binding, phosphatidylinositol-4,5-bisphosphate binding, phosphatidylinositol-3,4,5-trisphosphate binding InterPro: V(D)J recombination-activating protein 1, RAG nonamer-binding domain, Zinc finger, C3HC4 RING-type, Zinc finger, RING-type, Zinc finger, RING/FYVE/PHD-type, Zinc finger, RING-type, ...InterPro: V(D)J recombination-activating protein 1, RAG nonamer-binding domain, Zinc finger, C3HC4 RING-type, Zinc finger, RING-type, Zinc finger, RING/FYVE/PHD-type, Zinc finger, RING-type, conserved site, Galactose oxidase/kelch, beta-propeller, Kelch-type beta propeller, V-D-J recombination activating protein 2, Recombination activating protein 2, PHD domain, Zinc finger, FYVE/PHD-type |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 150 mM NaCl, 20 mM HEPES, 5 mM MgCl2, 1 mM TCEP |
| Grid | Details: 400 mesh Quantifoil holey carbon grid, glow discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 120 K / Instrument: GATAN CRYOPLUNGE 3 / Method: Blot for 2.5 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 80 K / Max: 105 K / Average: 100 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 150,000 times magnification |
| Date | Nov 14, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Number real images: 150 / Average electron dose: 41 e/Å2 Details: Every image is the average of 30 frames recorded by the direct electron detector. Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 40410 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.0 Å / Resolution method: OTHER / Software - Name: SPIDER, Relion / Number images used: 12055 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)