[English] 日本語
 Yorodumi
Yorodumi- EMDB-6403: Infectious bursal disease virus polyprotein and large precursors ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6403 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Infectious bursal disease virus polyprotein and large precursors assemble into a procapsid that is matured by its embedded protease | |||||||||
 Map data Map data | Reconstruction of IBDV VLP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryo-electron microscopy / IBDV / procapsid / viral protease / protein processing / protein self-assembly / virus assembly / virus structure | |||||||||
| Biological species |   Infectious bursal disease virus (Gumboro virus) Infectious bursal disease virus (Gumboro virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 21.0 Å | |||||||||
 Authors Authors | Castrillo-Briceno M / Gomez-Blanco J / Monton M / Rueda P / Caston JR | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Infectious bursal disease virus polyprotein and large precursors assemble into a procapsid that is matured by its embedded protease Authors: Castrillo-Briceno M / Gomez-Blanco J / Monton M / Rueda P / Caston JR | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6403.map.gz emd_6403.map.gz | 33.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6403-v30.xml emd-6403-v30.xml emd-6403.xml emd-6403.xml | 8.4 KB 8.4 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6403.gif 400_6403.gif 80_6403.gif 80_6403.gif | 82.4 KB 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6403 http://ftp.pdbj.org/pub/emdb/structures/EMD-6403 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6403 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6403 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6403.map.gz / Format: CCP4 / Size: 35 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6403.map.gz / Format: CCP4 / Size: 35 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of IBDV VLP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.21 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
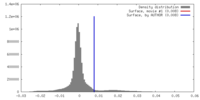
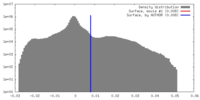
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : IBDV VLP
| Entire | Name: IBDV VLP |
|---|---|
| Components |
|
-Supramolecule #1000: IBDV VLP
| Supramolecule | Name: IBDV VLP / type: sample / ID: 1000 / Oligomeric state: 60 copies arranged into 20 trimers / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 2.8 MDa |
-Supramolecule #1: Infectious bursal disease virus
| Supramolecule | Name: Infectious bursal disease virus / type: virus / ID: 1 / NCBI-ID: 10995 / Sci species name: Infectious bursal disease virus / Sci species strain: Soroa / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  |
| Host system | Organism:  Trichoplusia ni (cabbage looper) / Recombinant cell: H5 Trichoplusia ni (cabbage looper) / Recombinant cell: H5 |
| Molecular weight | Theoretical: 2.8 MDa |
| Virus shell | Shell ID: 1 / Diameter: 700 Å / T number (triangulation number): 13 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.2 / Details: 150 mM NaCl, 20 mM CaCl2 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | May 1, 2011 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: each micrograph |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 21.0 Å / Resolution method: OTHER / Software - Name: Xmipp / Number images used: 424 |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)