[English] 日本語
 Yorodumi
Yorodumi- EMDB-63326: Cryo-EM structure of the histamine H1 receptor-Gi protein complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the histamine H1 receptor-Gi protein complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Class A GPCR / Histamine / G protein / Complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationHistamine receptors / histamine receptor activity / regulation of vascular permeability / cellular response to histamine / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma ...Histamine receptors / histamine receptor activity / regulation of vascular permeability / cellular response to histamine / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / G alpha (i) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / host cell mitochondrial membrane / host cell lipid droplet / G alpha (s) signalling events / photoreceptor outer segment membrane / G alpha (q) signalling events / symbiont-mediated transformation of host cell / spectrin binding / symbiont-mediated suppression of host TRAF-mediated signal transduction / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / alkylglycerophosphoethanolamine phosphodiesterase activity / symbiont-mediated perturbation of host cell cycle G1/S transition checkpoint / positive regulation of vasoconstriction / photoreceptor outer segment / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / cardiac muscle cell apoptotic process / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / photoreceptor inner segment / G protein-coupled serotonin receptor binding / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / bioluminescence / generation of precursor metabolites and energy / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / regulation of synaptic plasticity / visual learning / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / memory / ribonucleoside triphosphate phosphatase activity / ADP signalling through P2Y purinoceptor 12 / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / G alpha (z) signalling events / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / G-protein beta-subunit binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.23 Å | |||||||||
 Authors Authors | Matsuzaki Y / Sano FK / Oshima HS / Akasaka H / Kobayashi K / Tanaka T / Itoh Y / Shihoya W / Kise Y / Kusakizako T / Nureki O | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2025 Journal: Commun Biol / Year: 2025Title: Structural insights into ligand recognition and G protein preferences across histamine receptors. Authors: Yuma Matsuzaki / Fumiya K Sano / Hidetaka S Oshima / Hiroaki Akasaka / Kazuhiro Kobayashi / Tatsuki Tanaka / Yuzuru Itoh / Wataru Shihoya / Yoshiaki Kise / Tsukasa Kusakizako / Asuka Inoue / Osamu Nureki /  Abstract: Histamine exerts critical physiological roles by activating four receptor subtypes, each exhibiting a specific G protein preference. Among these, the histamine H receptor (HR) modulates chemotaxis ...Histamine exerts critical physiological roles by activating four receptor subtypes, each exhibiting a specific G protein preference. Among these, the histamine H receptor (HR) modulates chemotaxis and interferon production through G protein activation, suggesting its therapeutic potential. Despite its physiological significance, the mechanisms underlying HR signalling and G protein preference across histamine receptors remain poorly understood. Here, we present the cryo-electron microscopy structure of the HR-G complex, revealing unique mechanisms of histamine recognition and receptor activation. We further solved the structures of the histamine H receptor (HR) bound to the non-canonical G proteins G and G. Through a combination of functional and computational analyses, we identified the intracellular loop 2 as a critical determinant of G protein preference in HR and HR. Collectively, our comprehensive study revealed the structural basis for distinct mechanisms of ligand recognition and receptor activation, offering a profound insight into G protein preference across receptor subtypes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_63326.map.gz emd_63326.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-63326-v30.xml emd-63326-v30.xml emd-63326.xml emd-63326.xml | 25.1 KB 25.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_63326.png emd_63326.png | 110.6 KB | ||
| Masks |  emd_63326_msk_1.map emd_63326_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-63326.cif.gz emd-63326.cif.gz | 7.5 KB | ||
| Others |  emd_63326_half_map_1.map.gz emd_63326_half_map_1.map.gz emd_63326_half_map_2.map.gz emd_63326_half_map_2.map.gz | 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-63326 http://ftp.pdbj.org/pub/emdb/structures/EMD-63326 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63326 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63326 | HTTPS FTP |
-Related structure data
| Related structure data |  9lrdMC  9lrbC  9lrcC  9lreC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_63326.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_63326.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.29111 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_63326_msk_1.map emd_63326_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_63326_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_63326_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Histamine H1 receptor-Gi protein complex
| Entire | Name: Histamine H1 receptor-Gi protein complex |
|---|---|
| Components |
|
-Supramolecule #1: Histamine H1 receptor-Gi protein complex
| Supramolecule | Name: Histamine H1 receptor-Gi protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.415031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGGQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.744371 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.547685 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.720795 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAAASSEDL YFQ |
-Macromolecule #5: Histamine H1 receptor,Genome polyprotein
| Macromolecule | Name: Histamine H1 receptor,Genome polyprotein / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 90.333703 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKEFMSLP NSSCLLEDKM CEGNKTTMAS PQLMPLVVVL STICLVTVGL NLLVLYAVRS ERKLHTVGN LYIVSLSVAD LIVGAVVMPM NILYLLMSKW SLGRPLCLFW LSMDYVASTA SIFSVFILCI DRYRSVQQPL R YLKYRTKT ...String: MKTIIALSYI FCLVFADYKD DDDKEFMSLP NSSCLLEDKM CEGNKTTMAS PQLMPLVVVL STICLVTVGL NLLVLYAVRS ERKLHTVGN LYIVSLSVAD LIVGAVVMPM NILYLLMSKW SLGRPLCLFW LSMDYVASTA SIFSVFILCI DRYRSVQQPL R YLKYRTKT RASATILGAW FLSFLWVIPI LGWNHFMQQT SVRREDKCET DFYDVTWFKV MTAIINFYLP TLLMLWFYAK IY KAVRQHC QHRELINRSL PSFSEIKLRP ENPKGDAKKP GKESPWEVLK RKPKDAGGGS VLKSPSQTPK EMKSPVVFSQ EDD REVDKL YCFPLDIVHM QAAAEGSSRD YVAVNRSHGQ LKTDEQGLNT HGASEISEDQ MLGDSQSFSR TDSDTTTETA PGKG KLRSG SNTGLDYIKF TWKRLRSHSR QYVSGLHMNR ERKAAKQLGF IMAAFILCWI PYFIFFMVIA FCKNCCNEHL HMFTI WLGY INSTLNPLIY PLCNENFKKT FKRILHIRSG GSGGGGSGGS SSGGGSSGGS GGGSGGSGGL EVLFQGPVSK GEELFT GVV PILVELDGDV NGHKFSVSGE GEGDATYGKL TLKFICTTGK LPVPWPTLVT TLTYGVQCFS RYPDHMKQHD FFKSAMP EG YVQERTIFFK DDGNYKTRAE VKFEGDTLVN RIELKGIDFK EDGNILGHKL EYNYNSHNVY IMADKQKNGI KVNFKIRH N IEDGSVQLAD HYQQNTPIGD GPVLLPDNHY LSTQSKLSKD PNEKRDHMVL LEFVTAAGIT LGMDELYKSG LRSHHHHHH HH UniProtKB: Histamine H1 receptor, Genome polyprotein |
-Macromolecule #6: HISTAMINE
| Macromolecule | Name: HISTAMINE / type: ligand / ID: 6 / Number of copies: 1 / Formula: HSM |
|---|---|
| Molecular weight | Theoretical: 111.145 Da |
| Chemical component information | 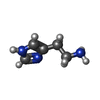 ChemComp-HSM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 10248 / Average electron dose: 48.96 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































