+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6310 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A Virus that Infects a Hyperthermophile Encapsidates A-Form DNA | |||||||||
 Map data Map data | Reconstruction of SIRV2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | archaeal virus / helical symmetry | |||||||||
| Function / homology | Sulfolobus virus, coat protein, C-terminal / Sulfolobus virus coat protein C terminal / helical viral capsid / DNA binding / Major capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |   Sulfolobus islandicus rod-shaped virus 2 Sulfolobus islandicus rod-shaped virus 2 | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | DiMaio F / Yu X / Rensen E / Krupovic M / Prangishvili D / Egelman E | |||||||||
 Citation Citation |  Journal: Science / Year: 2015 Journal: Science / Year: 2015Title: Virology. A virus that infects a hyperthermophile encapsidates A-form DNA. Authors: Frank DiMaio / Xiong Yu / Elena Rensen / Mart Krupovic / David Prangishvili / Edward H Egelman /   Abstract: Extremophiles, microorganisms thriving in extreme environmental conditions, must have proteins and nucleic acids that are stable at extremes of temperature and pH. The nonenveloped, rod-shaped virus ...Extremophiles, microorganisms thriving in extreme environmental conditions, must have proteins and nucleic acids that are stable at extremes of temperature and pH. The nonenveloped, rod-shaped virus SIRV2 (Sulfolobus islandicus rod-shaped virus 2) infects the hyperthermophilic acidophile Sulfolobus islandicus, which lives at 80°C and pH 3. We have used cryo-electron microscopy to generate a three-dimensional reconstruction of the SIRV2 virion at ~4 angstrom resolution, which revealed a previously unknown form of virion organization. Although almost half of the capsid protein is unstructured in solution, this unstructured region folds in the virion into a single extended α helix that wraps around the DNA. The DNA is entirely in the A-form, which suggests a common mechanism with bacterial spores for protecting DNA in the most adverse environments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6310.map.gz emd_6310.map.gz | 29.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6310-v30.xml emd-6310-v30.xml emd-6310.xml emd-6310.xml | 8.5 KB 8.5 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6310.gif 400_6310.gif 80_6310.gif 80_6310.gif | 76.7 KB 5.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6310 http://ftp.pdbj.org/pub/emdb/structures/EMD-6310 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6310 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6310 | HTTPS FTP |
-Related structure data
| Related structure data |  3j9xMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10060 (Title: Subset of image stack used for 3D reconstruction / Data size: 35.9 EMPIAR-10060 (Title: Subset of image stack used for 3D reconstruction / Data size: 35.9 Data #1: image segments of SIRV2 [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6310.map.gz / Format: CCP4 / Size: 31.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6310.map.gz / Format: CCP4 / Size: 31.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of SIRV2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
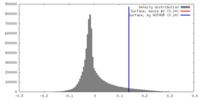
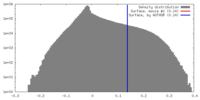
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SIRV2
| Entire | Name: SIRV2 |
|---|---|
| Components |
|
-Supramolecule #1000: SIRV2
| Supramolecule | Name: SIRV2 / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Sulfolobus islandicus rod-shaped virus 2
| Supramolecule | Name: Sulfolobus islandicus rod-shaped virus 2 / type: virus / ID: 1 / Name.synonym: SIRV2 / NCBI-ID: 157899 / Sci species name: Sulfolobus islandicus rod-shaped virus 2 / Database: NCBI / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No / Syn species name: SIRV2 |
|---|---|
| Host (natural) | Organism:   Sulfolobus islandicus (archaea) / synonym: ARCHAEA Sulfolobus islandicus (archaea) / synonym: ARCHAEA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Sep 19, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 437 / Average electron dose: 20 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | IHRSR |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 2.9 Å Applied symmetry - Helical parameters - Δ&Phi: 24.5 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: OTHER / Software - Name: Spider |
| CTF correction | Details: multiply images by CTF, divide reconstruction by sum of CTF**2 |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)