[English] 日本語
 Yorodumi
Yorodumi- EMDB-6291: 16 Angstrom cryo-EM reconstruction of the alpha, beta, gamma TF55... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6291 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 16 Angstrom cryo-EM reconstruction of the alpha, beta, gamma TF55 chaperonin | |||||||||
 Map data Map data | Cryo-EM reconstruction of the Sulfolobus solfataricus TF55 alpha/beta/gamma chaperonin | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TF55 chaperonin / protein folding / Sulfolobus solfataricus / cryo-EM | |||||||||
| Biological species |   Sulfolobus solfataricus (archaea) Sulfolobus solfataricus (archaea) | |||||||||
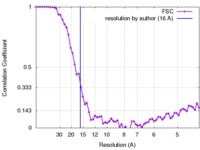
| Method | single particle reconstruction / cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Chaston JJ / Stewart AG / Smits C / Aragao D / Struwe W / Benesch J / Xwong A / Ling M / Ashsan B / Sandin S ...Chaston JJ / Stewart AG / Smits C / Aragao D / Struwe W / Benesch J / Xwong A / Ling M / Ashsan B / Sandin S / Rhodes D / Molugu SK / Bernal RA / Stock D | |||||||||
 Citation Citation |  Journal: Structure / Year: 2016 Journal: Structure / Year: 2016Title: Structural and Functional Insights into the Evolution and Stress Adaptation of Type II Chaperonins. Authors: Jessica J Chaston / Callum Smits / David Aragão / Andrew S W Wong / Bilal Ahsan / Sara Sandin / Sudheer K Molugu / Sanjay K Molugu / Ricardo A Bernal / Daniela Stock / Alastair G Stewart /    Abstract: Chaperonins are essential biological complexes assisting protein folding in all kingdoms of life. Whereas homooligomeric bacterial GroEL binds hydrophobic substrates non-specifically, the ...Chaperonins are essential biological complexes assisting protein folding in all kingdoms of life. Whereas homooligomeric bacterial GroEL binds hydrophobic substrates non-specifically, the heterooligomeric eukaryotic CCT binds specifically to distinct classes of substrates. Sulfolobales, which survive in a wide range of temperatures, have evolved three different chaperonin subunits (α, β, γ) that form three distinct complexes tailored for different substrate classes at cold, normal, and elevated temperatures. The larger octadecameric β complexes cater for substrates under heat stress, whereas smaller hexadecameric αβ complexes prevail under normal conditions. The cold-shock complex contains all three subunits, consistent with greater substrate specificity. Structural analysis using crystallography and electron microscopy reveals the geometry of these complexes and shows a novel arrangement of the α and β subunits in the hexadecamer enabling incorporation of the γ subunit. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6291.map.gz emd_6291.map.gz | 3.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6291-v30.xml emd-6291-v30.xml emd-6291.xml emd-6291.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6291_fsc.xml emd_6291_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_6291.jpg emd_6291.jpg | 824.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6291 http://ftp.pdbj.org/pub/emdb/structures/EMD-6291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6291 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6291.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6291.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the Sulfolobus solfataricus TF55 alpha/beta/gamma chaperonin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.174 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
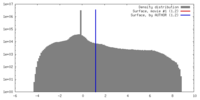
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cryo-EM reconstruction of the TF55 chaperonin containing the alph...
| Entire | Name: Cryo-EM reconstruction of the TF55 chaperonin containing the alpha, beta and gamma subunits |
|---|---|
| Components |
|
-Supramolecule #1000: Cryo-EM reconstruction of the TF55 chaperonin containing the alph...
| Supramolecule | Name: Cryo-EM reconstruction of the TF55 chaperonin containing the alpha, beta and gamma subunits type: sample / ID: 1000 Oligomeric state: Octadecamer composed of 6 alpha, 6 beta, and 6 gamma subunits per complex Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 1.08 MDa / Method: sequence |
-Macromolecule #1: TF55 chaperonin
| Macromolecule | Name: TF55 chaperonin / type: protein_or_peptide / ID: 1 Details: The TF55 chaperonin is composed of three proteins (alpha, beta, and gamma) arranged in an alternating fashion to form a 9-subunit ring (3 of each protein) which then stacks back-to-back with ...Details: The TF55 chaperonin is composed of three proteins (alpha, beta, and gamma) arranged in an alternating fashion to form a 9-subunit ring (3 of each protein) which then stacks back-to-back with a second ring, forming an 18-subunit complex. Number of copies: 18 / Oligomeric state: octadecamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Sulfolobus solfataricus (archaea) / Strain: DSM1617 Sulfolobus solfataricus (archaea) / Strain: DSM1617 |
| Molecular weight | Theoretical: 1 MDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM Tris-Cl, pH 8.0, 2 mM MgCl2, 1 mM EDTA, 50 mM NaCl |
| Grid | Details: 400-mesh copper grid with holey carbon film support (Quantifoil R2/2) |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 108 K / Instrument: HOMEMADE PLUNGER Method: 3 microliters of sample were blotted off the Quantifoil R2/2 grid for 3 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FS |
|---|---|
| Temperature | Min: 83 K / Max: 103 K / Average: 93 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 250,000 times magnification. |
| Specialist optics | Energy filter - Name: JEOL in-column / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Date | Jun 20, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Sampling interval: 15 µm / Number real images: 16534 / Average electron dose: 25 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 69000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder: 626 Gatan holder / Specimen holder model: GATAN LIQUID NITROGEN |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)