[English] 日本語
 Yorodumi
Yorodumi- EMDB-62898: Structure of Western equine encephalitis virus McMillan strain in... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Western equine encephalitis virus McMillan strain in complex with VLDLR LA1-2 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Western equine encephalitis virus / WEEV / receptor / complex / VLDLR / glycoprotein / VIRAL PROTEIN / McMillan strain / LA1-2 | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationreelin receptor activity / VLDL clearance / glycoprotein transport / very-low-density lipoprotein particle receptor activity / ventral spinal cord development / Reelin signalling pathway / very-low-density lipoprotein particle binding / low-density lipoprotein particle receptor activity / very-low-density lipoprotein particle clearance / togavirin ...reelin receptor activity / VLDL clearance / glycoprotein transport / very-low-density lipoprotein particle receptor activity / ventral spinal cord development / Reelin signalling pathway / very-low-density lipoprotein particle binding / low-density lipoprotein particle receptor activity / very-low-density lipoprotein particle clearance / togavirin / reelin-mediated signaling pathway / very-low-density lipoprotein particle / positive regulation of dendrite development / cargo receptor activity / T=4 icosahedral viral capsid / lipid transport / dendrite morphogenesis / regulation of synapse assembly / apolipoprotein binding / cholesterol metabolic process / clathrin-coated pit / receptor-mediated endocytosis / VLDLR internalisation and degradation / memory / calcium-dependent protein binding / nervous system development / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / receptor complex / symbiont-mediated suppression of host gene expression / lysosomal membrane / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / calcium ion binding / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane / glutamatergic synapse / virion membrane / structural molecule activity / signal transduction / proteolysis / RNA binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Western equine encephalitis virus / Western equine encephalitis virus /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||
 Authors Authors | Ma B / Cao Z / Ding W / Zhang X / Xiang Y / Cao D | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2025 Journal: Cell Rep / Year: 2025Title: Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus. Authors: Bingting Ma / Ziyi Cao / Weijia Ding / Xinzheng Zhang / Ye Xiang / Duanfang Cao /  Abstract: Western equine encephalitis virus (WEEV) enters cells via various receptors. Here, we report the cryoelectron microscopy (cryo-EM) structures of WEEV in complex with its receptors PCDH10 and very-low- ...Western equine encephalitis virus (WEEV) enters cells via various receptors. Here, we report the cryoelectron microscopy (cryo-EM) structures of WEEV in complex with its receptors PCDH10 and very-low-density lipoprotein receptor (VLDLR). Structural analysis shows that PCDH10 binds in the cleft formed by adjacent E2-E1 heterodimers of WEEV through its EC1 ectodomain. Residues of viral envelope proteins involved in the interactions with PCDH10 EC1 are unique to WEEV. The strain-specific receptor VLDLR binds WEEV strain McMillan through two consecutive ecto-LDLR class A (LA) repeats. LA1-2, LA2-3, LA3-4, LA4-5, and LA5-6 of VLDLR all have detectable interactions with WEEV. Detailed structures of WEEV in complex with LA1-2 and LA2-3 show that the N-terminal LA repeat binds in the cleft and that the C-terminal LA repeat is attached to the E2 B domain. The acquisition of a single E2 mutation (V265F) allows WEEV strain 71V-1658, originally unable to bind VLDLR, to gain this receptor-binding ability. The binding of VLDLR to WEEV is in a mode different from those of other alphaviruses. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_62898.map.gz emd_62898.map.gz | 2.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-62898-v30.xml emd-62898-v30.xml emd-62898.xml emd-62898.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_62898.png emd_62898.png | 87.3 KB | ||
| Masks |  emd_62898_msk_1.map emd_62898_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-62898.cif.gz emd-62898.cif.gz | 6.2 KB | ||
| Others |  emd_62898_half_map_1.map.gz emd_62898_half_map_1.map.gz emd_62898_half_map_2.map.gz emd_62898_half_map_2.map.gz | 23.2 MB 23.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-62898 http://ftp.pdbj.org/pub/emdb/structures/EMD-62898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62898 | HTTPS FTP |
-Validation report
| Summary document |  emd_62898_validation.pdf.gz emd_62898_validation.pdf.gz | 757 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_62898_full_validation.pdf.gz emd_62898_full_validation.pdf.gz | 756.3 KB | Display | |
| Data in XML |  emd_62898_validation.xml.gz emd_62898_validation.xml.gz | 10.7 KB | Display | |
| Data in CIF |  emd_62898_validation.cif.gz emd_62898_validation.cif.gz | 12.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62898 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62898 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62898 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62898 | HTTPS FTP |
-Related structure data
| Related structure data |  9l99MC  9l1nC  9l9aC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_62898.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_62898.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_62898_msk_1.map emd_62898_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
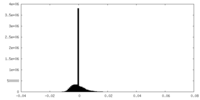
| Density Histograms |
-Half map: #1
| File | emd_62898_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
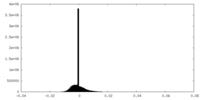
| Density Histograms |
-Half map: #2
| File | emd_62898_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : WEEV McMillan strain in complex with its receptor VLDLR LA1-2
| Entire | Name: WEEV McMillan strain in complex with its receptor VLDLR LA1-2 |
|---|---|
| Components |
|
-Supramolecule #1: WEEV McMillan strain in complex with its receptor VLDLR LA1-2
| Supramolecule | Name: WEEV McMillan strain in complex with its receptor VLDLR LA1-2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Western equine encephalitis virus / Strain: McMillan strian Western equine encephalitis virus / Strain: McMillan strian |
-Macromolecule #1: Spike glycoprotein E1
| Macromolecule | Name: Spike glycoprotein E1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Western equine encephalitis virus Western equine encephalitis virus |
| Molecular weight | Theoretical: 44.468277 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: FEHATTVPNV PGIPYKALVE RAGYAPLNLE ITVVSSELTP STNKEYVTCK FHTVIPSPQV KCCGSLECKA SSKADYTCRV FGGVYPFMW GGAQCFCDSE NTQLSEAYVE FAPDCTIDHA VALKVHTAAL KVGLRIVYGN TTAHLDTFVN GVTPGSSRDL K VIAGPISA ...String: FEHATTVPNV PGIPYKALVE RAGYAPLNLE ITVVSSELTP STNKEYVTCK FHTVIPSPQV KCCGSLECKA SSKADYTCRV FGGVYPFMW GGAQCFCDSE NTQLSEAYVE FAPDCTIDHA VALKVHTAAL KVGLRIVYGN TTAHLDTFVN GVTPGSSRDL K VIAGPISA AFSPFDHKVV IRKGLVYNYD FPEYGAMKPG AFGDIQASSL DATDIVARTD IRLLKPSVKN IHVPYTQAVS GY EMWKNNS GRPLQETAPF GCKIEVEPLR ASNCAYGHIP ISIDIPDAAF VRSSESPTIL EVSCTVADCI YSADFGGSLT LQY KADREG HCPVHSHSTT AVLKEATTHV TATGSITLHF STSSPQANFI VSLCGKKTTC NAECKPPADH IIGEPHKVDQ EFQA AVSKT SWNWLL UniProtKB: Structural polyprotein |
-Macromolecule #2: Spike glycoprotein E2
| Macromolecule | Name: Spike glycoprotein E2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Western equine encephalitis virus Western equine encephalitis virus |
| Molecular weight | Theoretical: 41.635254 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ITDDFTLTSP YLGFCPYCRH SAPCFSPIKI ENVWDESDDG SIRIQVSAQF GYNQAGTADV TKFRYMSYDH DHDIKEDSME KIAISTSGP CRRLGHKGYF LLAQCPPGDS VTVSITSGAS ENSCTVEKKI RRKFVGREEY LFPPVQGKLV KCHVYDRLKE T SAGYITMH ...String: ITDDFTLTSP YLGFCPYCRH SAPCFSPIKI ENVWDESDDG SIRIQVSAQF GYNQAGTADV TKFRYMSYDH DHDIKEDSME KIAISTSGP CRRLGHKGYF LLAQCPPGDS VTVSITSGAS ENSCTVEKKI RRKFVGREEY LFPPVQGKLV KCHVYDRLKE T SAGYITMH RPGPHAYKSY LKEASGEVYI KPPSGKNVTY ECKCGDYSTG IVSTQTKMNG CTKARQCIAY KLDQTKWVFN SP DLIRHTD HSVQGKLHIP FRLTPTVCPV PLAHTPTVTK WFKGITLHLT ATRPTLLTTR KLGLRADATA EWITGTTSRN FSV GREGLE YVWGNHEPVR VWAQESAPGD PHGWPHEIII HYYHRHPVYT VIV UniProtKB: Structural polyprotein |
-Macromolecule #3: Very low-density lipoprotein receptor
| Macromolecule | Name: Very low-density lipoprotein receptor / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.854691 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AKCEPSQFQC TNGRCITLLW KCDGDEDCVD GSDEKNCVKK TCAESDFVCN NGQCVPSRWK CDGDPDCEDG SDESPEQCHM UniProtKB: Very low-density lipoprotein receptor |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)