+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6252 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | VRC01 Fab in complex with BG505 SOSIP.664 Trimer | |||||||||
 Map data Map data | VRC01 Fab in complex with BG505 SOSIP.664 Trimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 / Fab / Env trimer | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 22.0 Å | |||||||||
 Authors Authors | Torres JL / Ozorowski G / Derking R / Sanders RW / Ward AB | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2015 Journal: PLoS Pathog / Year: 2015Title: Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. Authors: Ronald Derking / Gabriel Ozorowski / Kwinten Sliepen / Anila Yasmeen / Albert Cupo / Jonathan L Torres / Jean-Philippe Julien / Jeong Hyun Lee / Thijs van Montfort / Steven W de Taeye / Mark ...Authors: Ronald Derking / Gabriel Ozorowski / Kwinten Sliepen / Anila Yasmeen / Albert Cupo / Jonathan L Torres / Jean-Philippe Julien / Jeong Hyun Lee / Thijs van Montfort / Steven W de Taeye / Mark Connors / Dennis R Burton / Ian A Wilson / Per-Johan Klasse / Andrew B Ward / John P Moore / Rogier W Sanders /    Abstract: The trimeric envelope (Env) spike is the focus of vaccine design efforts aimed at generating broadly neutralizing antibodies (bNAbs) to protect against HIV-1 infection. Three recent developments have ...The trimeric envelope (Env) spike is the focus of vaccine design efforts aimed at generating broadly neutralizing antibodies (bNAbs) to protect against HIV-1 infection. Three recent developments have facilitated a thorough investigation of the antigenic structure of the Env trimer: 1) the isolation of many bNAbs against multiple different epitopes; 2) the generation of a soluble trimer mimic, BG505 SOSIP.664 gp140, that expresses most bNAb epitopes; 3) facile binding assays involving the oriented immobilization of tagged trimers. Using these tools, we generated an antigenic map of the trimer by antibody cross-competition. Our analysis delineates three well-defined epitope clusters (CD4 binding site, quaternary V1V2 and Asn332-centered oligomannose patch) and new epitopes at the gp120-gp41 interface. It also identifies the relationships among these clusters. In addition to epitope overlap, we defined three more ways in which antibodies can cross-compete: steric competition from binding to proximal but non-overlapping epitopes (e.g., PGT151 inhibition of 8ANC195 binding); allosteric inhibition (e.g., PGT145 inhibition of 1NC9, 8ANC195, PGT151 and CD4 binding); and competition by reorientation of glycans (e.g., PGT135 inhibition of CD4bs bNAbs, and CD4bs bNAb inhibition of 8ANC195). We further demonstrate that bNAb binding can be complex, often affecting several other areas of the trimer surface beyond the epitope. This extensive analysis of the antigenic structure and the epitope interrelationships of the Env trimer should aid in design of both bNAb-based therapies and vaccines intended to induce bNAbs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6252.map.gz emd_6252.map.gz | 13.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6252-v30.xml emd-6252-v30.xml emd-6252.xml emd-6252.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6252.gif 400_6252.gif 80_6252.gif 80_6252.gif | 12.7 KB 1.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6252 http://ftp.pdbj.org/pub/emdb/structures/EMD-6252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6252 | HTTPS FTP |
-Validation report
| Summary document |  emd_6252_validation.pdf.gz emd_6252_validation.pdf.gz | 77.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6252_full_validation.pdf.gz emd_6252_full_validation.pdf.gz | 76.9 KB | Display | |
| Data in XML |  emd_6252_validation.xml.gz emd_6252_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6252 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6252 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6252 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6252 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6252.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6252.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VRC01 Fab in complex with BG505 SOSIP.664 Trimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Fab of VRC01 human monoclonal antibody bound to HIV-1 Env gp140 B...
| Entire | Name: Fab of VRC01 human monoclonal antibody bound to HIV-1 Env gp140 BG505 SOSIP.664 |
|---|---|
| Components |
|
-Supramolecule #1000: Fab of VRC01 human monoclonal antibody bound to HIV-1 Env gp140 B...
| Supramolecule | Name: Fab of VRC01 human monoclonal antibody bound to HIV-1 Env gp140 BG505 SOSIP.664 type: sample / ID: 1000 Details: Size-exclusion-chromatography-purified gp140 trimers were used. Oligomeric state: One Fab molecule binds to one BG505 SOSIP.664 gp140 monomer Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 570 KDa |
-Macromolecule #1: Soluble HIV-1 Envelope glycoprotein
| Macromolecule | Name: Soluble HIV-1 Envelope glycoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: SOSIP / Number of copies: 1 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: BG505 / synonym: HIV-1 Human immunodeficiency virus 1 / Strain: BG505 / synonym: HIV-1 |
| Molecular weight | Theoretical: 420 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F / Recombinant plasmid: pPPI4 Homo sapiens (human) / Recombinant cell: HEK293F / Recombinant plasmid: pPPI4 |
-Macromolecule #2: Fab of VRC01 human monoclonal antibody
| Macromolecule | Name: Fab of VRC01 human monoclonal antibody / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Oligomeric state: dimer (heavy chain + light chain) / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 50 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM Tris-HCl, pH 7.4, 150 mM NaCl |
| Staining | Type: NEGATIVE Details: 3 uL of protein was applied and blotted; then 2% w/v uranyl formate was applied for 45 seconds. |
| Grid | Details: 400 Cu mesh grid with carbon support, plasma-cleaned |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 52,000x magnification. |
| Date | Oct 27, 2014 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 174 / Average electron dose: 25 e/Å2 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle min: -50 |
- Image processing
Image processing
| Details | The particles were selected using an automatic selection program. Class averages with 3 Fabs bound to one gp140 trimer were placed into a substack and used for 3D reconstruction. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 22.0 Å / Resolution method: OTHER / Software - Name: EMAN2, SPARX / Number images used: 7861 |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera

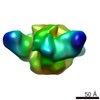

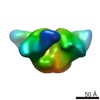

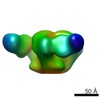



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















