+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of isw1-nucleosome complex in Apo state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Chromatin Remodeler / Nucleosome / DNA BINDING PROTEIN / DNA BINDING PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationIsw1b complex / Isw1a complex / Isw1 complex / regulation of transcriptional start site selection at RNA polymerase II promoter / nucleolar chromatin / negative regulation of RNA export from nucleus / negative regulation of IRE1-mediated unfolded protein response / regulation of chromatin organization / rDNA binding / DNA-templated transcription elongation ...Isw1b complex / Isw1a complex / Isw1 complex / regulation of transcriptional start site selection at RNA polymerase II promoter / nucleolar chromatin / negative regulation of RNA export from nucleus / negative regulation of IRE1-mediated unfolded protein response / regulation of chromatin organization / rDNA binding / DNA-templated transcription elongation / nucleosome array spacer activity / sister chromatid cohesion / termination of RNA polymerase II transcription / termination of RNA polymerase I transcription / nucleosome binding / mRNA 3'-UTR binding / helicase activity / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / structural constituent of chromatin / heterochromatin formation / nucleosome / nucleosome assembly / response to heat / transcription cis-regulatory region binding / chromatin remodeling / hydrolase activity / protein heterodimerization activity / chromatin binding / regulation of DNA-templated transcription / chromatin / positive regulation of transcription by RNA polymerase II / DNA binding / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Sia Y / Pan H / Chen Z | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2025 Journal: Science / Year: 2025Title: Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis. Authors: Youyang Sia / Han Pan / Kangjing Chen / Zhucheng Chen /  Abstract: Chromatin remodelers utilize the energy of adenosine triphosphate (ATP) hydrolysis to slide nucleosomes, regulating chromatin structure and gene activity in cells. In this work, we report structures ...Chromatin remodelers utilize the energy of adenosine triphosphate (ATP) hydrolysis to slide nucleosomes, regulating chromatin structure and gene activity in cells. In this work, we report structures of imitation switch (ISWI) bound to the nucleosome during active ATP hydrolysis and remodeling, revealing conformational transitions of the remodeling motor across the adenosine triphosphatase (ATPase) cycle. The DNA strands were distorted accordingly, showing one full base-pair bulge and a loss of histone contact at the site of motor binding in the adenosine diphosphate* (ADP*) and apo* (unbound) states. We also identified several important elements for regulation of the remodeling activity. Notably, an enzyme conformation exiting the remodeling cycle reveals a linker DNA-sensing brake mechanism. Together, our findings elucidate a multistate model of ISWI action, providing a comprehensive mechanism of DNA translocation and regulation underpinning chromatin remodeling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61632.map.gz emd_61632.map.gz | 32.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61632-v30.xml emd-61632-v30.xml emd-61632.xml emd-61632.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_61632_fsc.xml emd_61632_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_61632.png emd_61632.png | 50.6 KB | ||
| Masks |  emd_61632_msk_1.map emd_61632_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-61632.cif.gz emd-61632.cif.gz | 7.3 KB | ||
| Others |  emd_61632_half_map_1.map.gz emd_61632_half_map_1.map.gz emd_61632_half_map_2.map.gz emd_61632_half_map_2.map.gz | 59.3 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61632 http://ftp.pdbj.org/pub/emdb/structures/EMD-61632 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61632 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61632 | HTTPS FTP |
-Related structure data
| Related structure data |  9jnzMC  9jnpC  9jntC  9jnuC  9jnvC  9jnwC  9jnxC  9jo2C  9jo5C  9liuC  9lj2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61632.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61632.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0825 Å | ||||||||||||||||||||||||||||||||||||
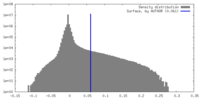
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_61632_msk_1.map emd_61632_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_61632_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61632_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of isw1-nucleosome complex in Apo state
| Entire | Name: Structure of isw1-nucleosome complex in Apo state |
|---|---|
| Components |
|
-Supramolecule #1: Structure of isw1-nucleosome complex in Apo state
| Supramolecule | Name: Structure of isw1-nucleosome complex in Apo state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Histone H3
| Macromolecule | Name: Histone H3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 15.30393 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGVKKPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQDFKT DLRFQSSAV MALQEASEAY LVALFEDTNL CAIHAKRVTI MPKDIQLARR IRGERA UniProtKB: Histone H3 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 11.263231 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKGGKGL GKGGAKRHRK VLRDNIQGIT KPAIRRLARR GGVKRISGLI YEETRGVLKV FLENVIRDAV TYTEHAKRKT VTAMDVVYA LKRQGRTLYG FGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.978241 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKQGGKT RAKAKTRSSR AGLQFPVGRV HRLLRKGNYA ERVGAGAPVY LAAVLEYLTA EILELAGNAA RDNKKTRIIP RHLQLAVRN DEELNKLLGR VTIAQGGVLP NIQSVLLPKK TESSKSAKSK UniProtKB: Histone H2A |
-Macromolecule #4: Histone H2B
| Macromolecule | Name: Histone H2B / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.524752 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AKSAPAPKKG SKKAVTKTQK KDGKKRRKTR KESYAIYVYK VLKQVHPDTG ISSKAMSIMN SFVNDVFERI AGEASRLAHY NKRSTITSR EIQTAVRLLL PGELAKHAVS EGTKAVTKYT SAK UniProtKB: Histone H2B |
-Macromolecule #7: ISWI chromatin-remodeling complex ATPase ISW1
| Macromolecule | Name: ISWI chromatin-remodeling complex ATPase ISW1 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 123.170516 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ENLKPFQVGL PPHDPESNKK RYLLKDANGK KFDLEGTTKR FEHLLSLSGL FKHFIESKAA KDPKFRQVLD VLEENKANGK GKGKHQDVR RRKTEHEEDA ELLKEEDSDD DESIEFQFRE SPAYVNGQLR PYQIQGVNWL VSLHKNKIAG ILADEMGLGK T LQTISFLG ...String: ENLKPFQVGL PPHDPESNKK RYLLKDANGK KFDLEGTTKR FEHLLSLSGL FKHFIESKAA KDPKFRQVLD VLEENKANGK GKGKHQDVR RRKTEHEEDA ELLKEEDSDD DESIEFQFRE SPAYVNGQLR PYQIQGVNWL VSLHKNKIAG ILADEMGLGK T LQTISFLG YLRYIEKIPG PFLVIAPKST LNNWLREINR WTPDVNAFIL QGDKEERAEL IQKKLLGCDF DVVIASYEII IR EKSPLKK INWEYIIIDE AHRIKNEESM LSQVLREFTS RNRLLITGTP LQNNLHELWA LLNFLLPDIF SDAQDFDDWF SSE STEEDQ DKIVKQLHTV LQPFLLRRIK SDVETSLLPK KELNLYVGMS SMQKKWYKKI LEKDLDAVNG SNGSKESKTR LLNI MMQLR KCCNHPYLFD GAEPGPPYTT DEHLVYNAAK LQVLDKLLKK LKEEGSRVLI FSQMSRLLDI LEDYCYFRNY EYCRI DGST AHEDRIQAID DYNAPDSKKF VFLLTTRAGG LGINLTSADV VVLYDSDWNP QADLQAMDRA HRIGQKKQVK VFRLVT DNS VEEKILERAT QKLRLDQLVI QQNRTSLKKK ENKADSKDAL LSMIQHGAAD VFKSGTSTGS AGTPEPGSGE KGDDIDL DE LLLKSENKTK SLNAKYETLG LDDLQKFNQD SAYEWNGQDF KKKIQRDIIS PLLLNPTKRE RKENYSIDNY YKDVLNTG R SSTPSHPRMP KPHVFHSHQL QPPQLKVLYE KERMWTAKKT GYVPTMDDVK AAYGDISDEE EKKQKLELLK LSVNNSQPL TEEEEKMKAD WESEGFTNWN KLEFRKFITV SGKYGRNSIQ AIARELAPGK TLEEVRAYAK AFWSNIERIE DYEKYLKIIE NEEEKIKRV KMQQEALRRK LSEYKNPFFD LKLKHPPSSN NKRTYSEEED RFILLMLFKY GLDRDDVYEL VRDEIRDCPL F ELDFYFRS RTPVELARRG NTLLQCLEKE FNAGIVLDDA TKDRMKKEDE NGKRIREEFA DQTANEKENV DGVESKKAKI ED TSNVGTE QLVAEKIPEN ETTH UniProtKB: ISWI chromatin-remodeling complex ATPase ISW1 |
-Macromolecule #5: DNA (146-MER)
| Macromolecule | Name: DNA (146-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44.825559 KDa |
| Sequence | String: (DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT)(DC) ...String: (DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT)(DC)(DT)(DA) (DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT)(DA) (DA)(DA) (DC)(DG)(DC)(DA)(DC)(DG)(DT) (DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT)(DC) (DC)(DC)(DC) (DC)(DG)(DC)(DG)(DT)(DT) (DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC)(DA) (DA)(DG)(DG)(DG) (DG)(DA)(DT)(DT)(DA) (DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT)(DC) (DT)(DC)(DC)(DA)(DG) (DG)(DC)(DA)(DC) (DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT)(DA) (DT)(DA)(DT)(DA)(DC)(DA) (DT)(DC)(DC) (DG)(DA)(DT) |
-Macromolecule #6: DNA (146-MER)
| Macromolecule | Name: DNA (146-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.305852 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC) ...String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT) (DA)(DA) (DA)(DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC) (DG)(DT)(DA) (DC)(DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC) (DA)(DA)(DT)(DT)(DG) (DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC) (DC)(DG)(DG)(DG)(DA)(DT) (DT)(DC)(DT) (DC)(DG)(DA) |
-Macromolecule #8: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 8 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)