[English] 日本語
 Yorodumi
Yorodumi- EMDB-6142: Cryo-EM reconstructions of E. coli ribosomal 30S subunit assembly... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6142 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstructions of E. coli ribosomal 30S subunit assembly intermediates | |||||||||
 Map data Map data | Reconstruction of Group I intermediate from cryo-EM analysis of affinity-purified 30S assembly intermediates (Frealign model 1 of 4) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome assembly / 30S subunit / Assembly intermediate | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 27.6 Å | |||||||||
 Authors Authors | Sashital DG / Greeman CA / Lyumkis D / Potter CS / Carragher B / Williamson JR | |||||||||
 Citation Citation |  Journal: Elife / Year: 2014 Journal: Elife / Year: 2014Title: A combined quantitative mass spectrometry and electron microscopy analysis of ribosomal 30S subunit assembly in E. coli. Authors: Dipali G Sashital / Candacia A Greeman / Dmitry Lyumkis / Clinton S Potter / Bridget Carragher / James R Williamson /  Abstract: Ribosome assembly is a complex process involving the folding and processing of ribosomal RNAs (rRNAs), concomitant binding of ribosomal proteins (r-proteins), and participation of numerous accessory ...Ribosome assembly is a complex process involving the folding and processing of ribosomal RNAs (rRNAs), concomitant binding of ribosomal proteins (r-proteins), and participation of numerous accessory cofactors. Here, we use a quantitative mass spectrometry/electron microscopy hybrid approach to determine the r-protein composition and conformation of 30S ribosome assembly intermediates in Escherichia coli. The relative timing of assembly of the 3' domain and the formation of the central pseudoknot (PK) structure depends on the presence of the assembly factor RimP. The central PK is unstable in the absence of RimP, resulting in the accumulation of intermediates in which the 3'-domain is unanchored and the 5'-domain is depleted for r-proteins S5 and S12 that contact the central PK. Our results reveal the importance of the cofactor RimP in central PK formation, and introduce a broadly applicable method for characterizing macromolecular assembly in cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6142.map.gz emd_6142.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6142-v30.xml emd-6142-v30.xml emd-6142.xml emd-6142.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6142.gif 400_6142.gif 80_6142.gif 80_6142.gif | 16.3 KB 1.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6142 http://ftp.pdbj.org/pub/emdb/structures/EMD-6142 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6142 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6142 | HTTPS FTP |
-Validation report
| Summary document |  emd_6142_validation.pdf.gz emd_6142_validation.pdf.gz | 78.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6142_full_validation.pdf.gz emd_6142_full_validation.pdf.gz | 77.6 KB | Display | |
| Data in XML |  emd_6142_validation.xml.gz emd_6142_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6142 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6142 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6142 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6142 | HTTPS FTP |
-Related structure data
| Related structure data |  6125C  6126C  6127C  6128C  6129C  6130C  6131C  6132C  6133C  6134C  6135C  6136C  6137C  6138C  6139C  6140C  6141C  6143C  6144C  6145C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6142.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6142.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of Group I intermediate from cryo-EM analysis of affinity-purified 30S assembly intermediates (Frealign model 1 of 4) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
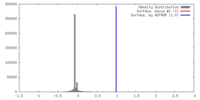
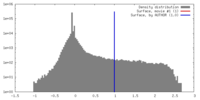
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Early (Group I) 30S ribosomal subunit assembly intermediate, class 1
| Entire | Name: Early (Group I) 30S ribosomal subunit assembly intermediate, class 1 |
|---|---|
| Components |
|
-Supramolecule #1000: Early (Group I) 30S ribosomal subunit assembly intermediate, class 1
| Supramolecule | Name: Early (Group I) 30S ribosomal subunit assembly intermediate, class 1 type: sample / ID: 1000 Details: Group I particles from early 30S assembly intermediates affinity-purified from E. coli rimP deletion strain Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 500 KDa / Method: Calculation of MW of known components |
-Supramolecule #1: 30S assembly intermediate
| Supramolecule | Name: 30S assembly intermediate / type: complex / ID: 1 / Name.synonym: 30S ribosomal subunit / Recombinant expression: No / Database: NCBI / Ribosome-details: ribosome-prokaryote: SSU 30S, PSR16s |
|---|---|
| Ref GO | 0: GO:0005840 |
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 500 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM Tris, pH 7.5, 100 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, 6 mM 2-mercaptoethanol |
| Grid | Details: C-flat grids (Protochips) with 2 micron diameter holes coated with a thin (2 nm) layer of continuous carbon support, plasma-cleaned for 5s |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 120 K / Instrument: GATAN CRYOPLUNGE 3 Method: Sample was applied to grid for 1 minute, then blotted for 3 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 80 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected using a live image of the power spectrum. |
| Date | Aug 29, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Number real images: 1253 / Average electron dose: 33.67 e/Å2 Details: The dose was fractionated over 30 frames (200 ms each) and image frames were aligned using a program generously provided by Yifan Cheng and Xueming Li. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 29000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Image frame sets (30 frames, 200 ms each) were aligned prior to image processing. Particles were selected using DoG picker in Appion. Group I particles were identified following extensive alignment and classification. |
|---|---|
| CTF correction | Details: Each micrograph |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 27.6 Å / Resolution method: OTHER / Software - Name: Xmipp, Frealign Details: Group I particles were sorted into 4 groups and reconstructed using Frealign 9. Number images used: 3407 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)