+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6123 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical nanotube formed from a 29-residue peptide | |||||||||
 Map data Map data | reconstruction of Form I polymer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | IHRSR / polymorphism / coiled-coils | |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Egelman EH / Xu C / DiMaio F / Magnotti E / Modlin C / Yu X / Wright E / Baker D / Conticello VP | |||||||||
 Citation Citation |  Journal: Structure / Year: 2015 Journal: Structure / Year: 2015Title: Structural plasticity of helical nanotubes based on coiled-coil assemblies. Authors: E H Egelman / C Xu / F DiMaio / E Magnotti / C Modlin / X Yu / E Wright / D Baker / V P Conticello /  Abstract: Numerous instances can be seen in evolution in which protein quaternary structures have diverged while the sequences of the building blocks have remained fairly conserved. However, the path through ...Numerous instances can be seen in evolution in which protein quaternary structures have diverged while the sequences of the building blocks have remained fairly conserved. However, the path through which such divergence has taken place is usually not known. We have designed two synthetic 29-residue α-helical peptides, based on the coiled-coil structural motif, that spontaneously self-assemble into helical nanotubes in vitro. Using electron cryomicroscopy with a newly available direct electron detection capability, we can achieve near-atomic resolution of these thin structures. We show how conservative changes of only one or two amino acids result in dramatic changes in quaternary structure, in which the assemblies can be switched between two very different forms. This system provides a framework for understanding how small sequence changes in evolution can translate into very large changes in supramolecular structure, a phenomenon that may have significant implications for the de novo design of synthetic peptide assemblies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6123.map.gz emd_6123.map.gz | 345.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6123-v30.xml emd-6123-v30.xml emd-6123.xml emd-6123.xml | 8.4 KB 8.4 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6123.gif 400_6123.gif 80_6123.gif 80_6123.gif | 51.2 KB 4.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6123 http://ftp.pdbj.org/pub/emdb/structures/EMD-6123 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6123 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6123 | HTTPS FTP |
-Related structure data
| Related structure data |  3j89MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6123.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6123.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | reconstruction of Form I polymer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
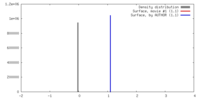
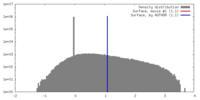
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : synthetic peptide
| Entire | Name: synthetic peptide (others) |
|---|---|
| Components |
|
-Supramolecule #1000: synthetic peptide
| Supramolecule | Name: synthetic peptide / type: sample / ID: 1000 / Oligomeric state: polymer / Number unique components: 1 |
|---|
-Macromolecule #1: synthetic peptide
| Macromolecule | Name: synthetic peptide / type: protein_or_peptide / ID: 1 / Details: QARILEADAEILRAYARILEAHAEILRAQ / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: FEI VITROBOT MARK IV |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Aug 1, 2013 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 260 / Average electron dose: 10 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.4 µm / Nominal defocus min: 1.4 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | IHRSR |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 2.2 Å Applied symmetry - Helical parameters - Δ&Phi: 87.1 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: OTHER / Software - Name: Spider |
| CTF correction | Details: micrographs multiplied by CTF |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)