[English] 日本語
 Yorodumi
Yorodumi- EMDB-5910: Cryo-EM structure of high salt treated immature 30S ribosomal sub... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5910 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of high salt treated immature 30S ribosomal subunit from rsga and rbfa deleted E.coli strain | |||||||||
 Map data Map data | the map is normalized to N(0,1) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | 30S subunit assembly / RsgA / RbfA / 17S rRNA processing | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 18.3 Å | |||||||||
 Authors Authors | Yang Z / Guo Q / Goto S / Chen Y / Li N / Yan K / Zhang Y / Muto A / Deng H / Himeno H ...Yang Z / Guo Q / Goto S / Chen Y / Li N / Yan K / Zhang Y / Muto A / Deng H / Himeno H / Lei J / Gao N | |||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2014 Journal: Protein Cell / Year: 2014Title: Structural insights into the assembly of the 30S ribosomal subunit in vivo: functional role of S5 and location of the 17S rRNA precursor sequence. Authors: Zhixiu Yang / Qiang Guo / Simon Goto / Yuling Chen / Ningning Li / Kaige Yan / Yixiao Zhang / Akira Muto / Haiteng Deng / Hyouta Himeno / Jianlin Lei / Ning Gao /  Abstract: The in vivo assembly of ribosomal subunits is a highly complex process, with a tight coordination between protein assembly and rRNA maturation events, such as folding and processing of rRNA ...The in vivo assembly of ribosomal subunits is a highly complex process, with a tight coordination between protein assembly and rRNA maturation events, such as folding and processing of rRNA precursors, as well as modifications of selected bases. In the cell, a large number of factors are required to ensure the efficiency and fidelity of subunit production. Here we characterize the immature 30S subunits accumulated in a factor-null Escherichia coli strain (∆rsgA∆rbfA). The immature 30S subunits isolated with varying salt concentrations in the buffer system show interesting differences on both protein composition and structure. Specifically, intermediates derived under the two contrasting salt conditions (high and low) likely reflect two distinctive assembly stages, the relatively early and late stages of the 3' domain assembly, respectively. Detailed structural analysis demonstrates a mechanistic coupling between the maturation of the 5' end of the 17S rRNA and the assembly of the 30S head domain, and attributes a unique role of S5 in coordinating these two events. Furthermore, our structural results likely reveal the location of the unprocessed terminal sequences of the 17S rRNA, and suggest that the maturation events of the 17S rRNA could be employed as quality control mechanisms on subunit production and protein translation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5910.map.gz emd_5910.map.gz | 7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5910-v30.xml emd-5910-v30.xml emd-5910.xml emd-5910.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  400_5910.gif 400_5910.gif 80_5910.gif 80_5910.gif | 41.7 KB 3.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5910 http://ftp.pdbj.org/pub/emdb/structures/EMD-5910 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5910 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5910 | HTTPS FTP |
-Validation report
| Summary document |  emd_5910_validation.pdf.gz emd_5910_validation.pdf.gz | 79.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5910_full_validation.pdf.gz emd_5910_full_validation.pdf.gz | 78.5 KB | Display | |
| Data in XML |  emd_5910_validation.xml.gz emd_5910_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5910 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5910 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5910 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5910 | HTTPS FTP |
-Related structure data
| Related structure data |  5900C  5904C  5905C  5906C  5907C  5908C  5909C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5910.map.gz / Format: CCP4 / Size: 7.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5910.map.gz / Format: CCP4 / Size: 7.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | the map is normalized to N(0,1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
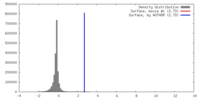
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cryo-EM structure of high salt treated immature 30S ribosomal sub...
| Entire | Name: Cryo-EM structure of high salt treated immature 30S ribosomal subunit from rsga and rbfa deleted E.coli strain |
|---|---|
| Components |
|
-Supramolecule #1000: Cryo-EM structure of high salt treated immature 30S ribosomal sub...
| Supramolecule | Name: Cryo-EM structure of high salt treated immature 30S ribosomal subunit from rsga and rbfa deleted E.coli strain type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 800 KDa / Theoretical: 800 KDa |
-Supramolecule #1: high salt treated immature 30S ribosomal subunit from rsga and rb...
| Supramolecule | Name: high salt treated immature 30S ribosomal subunit from rsga and rbfa deleted E.coli strain type: complex / ID: 1 / Name.synonym: immature 30S / Recombinant expression: No / Database: NCBI / Ribosome-details: ribosome-prokaryote: SSU 30S |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 800 KDa / Theoretical: 800 KDa |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Details: 10mM Tris-HCl, 60mM NH4Cl,10mM MgCl2, 0.1mM EDTA,1mM DTT |
|---|---|
| Staining | Type: NEGATIVE / Details: grids were prepared with an FEI Vitrobot Mark IV |
| Grid | Details: Quantifoil 2/4 grids were coated with carbon and glow discharged in a Harrick Plasma Cleaner for 30 seconds |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Method: blot for 1 second before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Nov 7, 2012 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (4k x 4k) / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.2 µm / Nominal defocus min: 0.77 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | this is a classification volume using RELION |
|---|---|
| CTF correction | Details: weiner filter |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 18.3 Å / Resolution method: OTHER / Software - Name: SPIDER / Number images used: 7536 |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)