+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5793 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A common solution to group 2 influenza virus neutralization | |||||||||
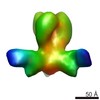
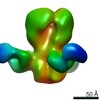
 Map data Map data | Influenza (A/Hong Kong/1/1968, H3N2) hemagglutinin bound to neutralizing antibody CR8043 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Antibody recognition / Hemagglutinin | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Influenza A virus Influenza A virus | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 21.0 Å | |||||||||
 Authors Authors | Friesen RHE / Leeb PS / Stoop EJM / Hoffman RMB / Ekiert DC / Bhabha G / Yu W / Juraszek J / Koudstaal W / Jongeneelen M ...Friesen RHE / Leeb PS / Stoop EJM / Hoffman RMB / Ekiert DC / Bhabha G / Yu W / Juraszek J / Koudstaal W / Jongeneelen M / Korse HJWM / Ophorst C / Brinkman-van der Linden ECM / Throsby M / Kwakkenbos MJ / Bakkerd AQ / Beaumont Y / Spits H / Kwaks T / Vogels R / Ward AB / Goudsmit J / Wilson IA | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2014 Journal: Proc Natl Acad Sci U S A / Year: 2014Title: A common solution to group 2 influenza virus neutralization. Authors: Robert H E Friesen / Peter S Lee / Esther J M Stoop / Ryan M B Hoffman / Damian C Ekiert / Gira Bhabha / Wenli Yu / Jarek Juraszek / Wouter Koudstaal / Mandy Jongeneelen / Hans J W M Korse / ...Authors: Robert H E Friesen / Peter S Lee / Esther J M Stoop / Ryan M B Hoffman / Damian C Ekiert / Gira Bhabha / Wenli Yu / Jarek Juraszek / Wouter Koudstaal / Mandy Jongeneelen / Hans J W M Korse / Carla Ophorst / Els C M Brinkman-van der Linden / Mark Throsby / Mark J Kwakkenbos / Arjen Q Bakker / Tim Beaumont / Hergen Spits / Ted Kwaks / Ronald Vogels / Andrew B Ward / Jaap Goudsmit / Ian A Wilson /  Abstract: The discovery and characterization of broadly neutralizing antibodies (bnAbs) against influenza viruses have raised hopes for the development of monoclonal antibody (mAb)-based immunotherapy and the ...The discovery and characterization of broadly neutralizing antibodies (bnAbs) against influenza viruses have raised hopes for the development of monoclonal antibody (mAb)-based immunotherapy and the design of universal influenza vaccines. Only one human bnAb (CR8020) specifically recognizing group 2 influenza A viruses has been previously characterized that binds to a highly conserved epitope at the base of the hemagglutinin (HA) stem and has neutralizing activity against H3, H7, and H10 viruses. Here, we report a second group 2 bnAb, CR8043, which was derived from a different germ-line gene encoding a highly divergent amino acid sequence. CR8043 has in vitro neutralizing activity against H3 and H10 viruses and protects mice against challenge with a lethal dose of H3N2 and H7N7 viruses. The crystal structure and EM reconstructions of the CR8043-H3 HA complex revealed that CR8043 binds to a site similar to the CR8020 epitope but uses an alternative angle of approach and a distinct set of interactions. The identification of another antibody against the group 2 stem epitope suggests that this conserved site of vulnerability has great potential for design of therapeutics and vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5793.map.gz emd_5793.map.gz | 25.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5793-v30.xml emd-5793-v30.xml emd-5793.xml emd-5793.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5793_1.jpg emd_5793_1.jpg | 55.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5793 http://ftp.pdbj.org/pub/emdb/structures/EMD-5793 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5793 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5793 | HTTPS FTP |
-Validation report
| Summary document |  emd_5793_validation.pdf.gz emd_5793_validation.pdf.gz | 79.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5793_full_validation.pdf.gz emd_5793_full_validation.pdf.gz | 78.4 KB | Display | |
| Data in XML |  emd_5793_validation.xml.gz emd_5793_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5793 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5793 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5793 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5793 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5793.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5793.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Influenza (A/Hong Kong/1/1968, H3N2) hemagglutinin bound to neutralizing antibody CR8043 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
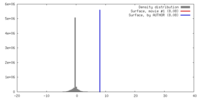
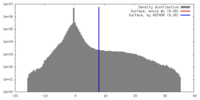
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Influenza (A/Hong Kong/1/1968, H3N2) hemagglutinin bound to neutr...
| Entire | Name: Influenza (A/Hong Kong/1/1968, H3N2) hemagglutinin bound to neutralizing antibody CR8043 |
|---|---|
| Components |
|
-Supramolecule #1000: Influenza (A/Hong Kong/1/1968, H3N2) hemagglutinin bound to neutr...
| Supramolecule | Name: Influenza (A/Hong Kong/1/1968, H3N2) hemagglutinin bound to neutralizing antibody CR8043 type: sample / ID: 1000 / Oligomeric state: One HA trimer bound to three Fabs / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 288 KDa |
-Macromolecule #1: CR8043 Fab
| Macromolecule | Name: CR8043 Fab / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 44 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
-Macromolecule #2: Influenza A/Hong Kong/1/1968 (H3N2) hemagglutinin
| Macromolecule | Name: Influenza A/Hong Kong/1/1968 (H3N2) hemagglutinin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Oligomeric state: trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Influenza A virus / Strain: A/Hong Kong/1/1968 Influenza A virus / Strain: A/Hong Kong/1/1968 |
| Molecular weight | Theoretical: 157 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: TBS |
| Staining | Type: NEGATIVE Details: Samples were applied to freshly glow-discharged grids and stained with 2% uranyl formate (20 seconds). |
| Grid | Details: 400 mesh copper with nitrocellulose and thin layer of carbon |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Temperature | Average: 293 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism corrected at 100,000 times magnification. |
| Date | May 18, 2013 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 243 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 55 |
- Image processing
Image processing
| Details | Projection matching seeded with a low-pass filtered hemagglutinin |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 21.0 Å / Resolution method: OTHER / Software - Name: Appion, Spider, Xmipp, Eman1, Sparx / Number images used: 10033 |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)