[English] 日本語
 Yorodumi
Yorodumi- EMDB-5701: Negative stained image reconstruction of HIV KNH11444 subtype A S... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5701 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.681.dV1V2V3 | |||||||||
 Map data Map data | Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.681.dV1V2V3 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Human immunodeficiency virus / antigen / vaccine development / neutralization / conformational change / micelles | |||||||||
| Biological species |   Human immunodeficiency virus Human immunodeficiency virus | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 17.0 Å | |||||||||
 Authors Authors | Khayat R / Lee JH / Wilson IA / Ward AB | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2013 Journal: J Virol / Year: 2013Title: Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. Authors: Per Johan Klasse / Rafael S Depetris / Robert Pejchal / Jean-Philippe Julien / Reza Khayat / Jeong Hyun Lee / Andre J Marozsan / Albert Cupo / Nicolette Cocco / Jacob Korzun / Anila Yasmeen ...Authors: Per Johan Klasse / Rafael S Depetris / Robert Pejchal / Jean-Philippe Julien / Reza Khayat / Jeong Hyun Lee / Andre J Marozsan / Albert Cupo / Nicolette Cocco / Jacob Korzun / Anila Yasmeen / Andrew B Ward / Ian A Wilson / Rogier W Sanders / John P Moore /  Abstract: We describe methods to improve the properties of soluble, cleaved gp140 trimers of the human immunodeficiency virus type 1 (HIV-1) envelope glycoproteins (Env) for use in structural studies and as ...We describe methods to improve the properties of soluble, cleaved gp140 trimers of the human immunodeficiency virus type 1 (HIV-1) envelope glycoproteins (Env) for use in structural studies and as immunogens. In the absence of nonionic detergents, gp140 of the KNH1144 genotype, terminating at residue 681 in gp41 (SOSIP.681), has a tendency to form higher-order complexes or aggregates, which is particularly undesirable for structure-based research. We found that this aggregation in the absence of detergent does not involve the V1, V2, or V3 variable regions of gp120. Moreover, we observed that detergent forms micelles around the membrane-proximal external region (MPER) of the SOSIP.681 gp140 trimers, whereas deletion of most of the MPER residues by terminating the gp140 at residue 664 (SOSIP.664) prevented the aggregation that otherwise occurs in SOSIP.681 in the absence of detergent. Although the MPER can contribute to trimer formation, truncation of most of it only modestly reduced trimerization and lacked global adverse effects on antigenicity. Thus, the MPER deletion minimally influenced the kinetics of the binding of soluble CD4 and a CD4-binding site antibody to immobilized trimers, as detected by surface plasmon resonance. Furthermore, the MPER deletion did not alter the overall three-dimensional structure of the trimers, as viewed by negative-stain electron microscopy. Homogeneous and aggregate-free MPER-truncated SOSIP Env trimers are therefore useful for immunogenicity and structural studies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5701.map.gz emd_5701.map.gz | 12.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5701-v30.xml emd-5701-v30.xml emd-5701.xml emd-5701.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5701.png emd_5701.png | 100.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5701 http://ftp.pdbj.org/pub/emdb/structures/EMD-5701 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5701 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5701 | HTTPS FTP |
-Validation report
| Summary document |  emd_5701_validation.pdf.gz emd_5701_validation.pdf.gz | 78.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5701_full_validation.pdf.gz emd_5701_full_validation.pdf.gz | 77.6 KB | Display | |
| Data in XML |  emd_5701_validation.xml.gz emd_5701_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5701 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5701 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5701 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5701 | HTTPS FTP |
-Related structure data
| Related structure data |  5700C  5702C  5703C  5704C  5705C  5706C 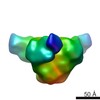 5707C  5708C  5709C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5701.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5701.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.681.dV1V2V3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.18 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
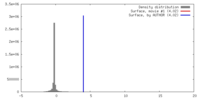
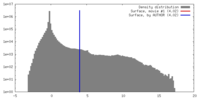
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HIV KNH11444 subtype A SOSIP.681.dV1V2V3
| Entire | Name: HIV KNH11444 subtype A SOSIP.681.dV1V2V3 |
|---|---|
| Components |
|
-Supramolecule #1000: HIV KNH11444 subtype A SOSIP.681.dV1V2V3
| Supramolecule | Name: HIV KNH11444 subtype A SOSIP.681.dV1V2V3 / type: sample / ID: 1000 / Details: The sample was monodisperse via SEC. / Oligomeric state: Trimer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 395 KDa |
-Macromolecule #1: Human immunodeficiency virus Envelope protein
| Macromolecule | Name: Human immunodeficiency virus Envelope protein / type: protein_or_peptide / ID: 1 / Name.synonym: gp120/gp41 / Number of copies: 3 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus / Strain: KNH1144 / synonym: HIV Human immunodeficiency virus / Strain: KNH1144 / synonym: HIV |
| Molecular weight | Theoretical: 395 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: 293S / Recombinant plasmid: pPI4 Homo sapiens (human) / Recombinant cell: 293S / Recombinant plasmid: pPI4 |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM HEPES, pH 7.5, 50 mM NaCl |
| Staining | Type: NEGATIVE / Details: 2% Uranyl Formate for 30 seconds |
| Grid | Details: 400 mesh copper grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 293 K / Max: 294 K / Average: 293 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification Legacy - Electron beam tilt params: -2 |
| Date | Oct 25, 2011 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (4k x 4k) / Digitization - Sampling interval: 10.9 µm / Number real images: 190 / Average electron dose: 16 e/Å2 / Details: Data collected on CCD / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 100000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 0.9 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 100000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 55 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)