[English] 日本語
 Yorodumi
Yorodumi- EMDB-5588: Electron cryo-microscopy of the yeast Mediator Cdk8 kinase module -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5588 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy of the yeast Mediator Cdk8 kinase module | |||||||||
 Map data Map data | Map of the CDK8 kinase module including Med12, Med13, Cdk8, and CycC | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mediator / kinase module / CDK8 / Med12 / Med13 / CycC | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 15.0 Å | |||||||||
 Authors Authors | Tsai KL / Sato S / Tomomori-Sato C / Conaway RC / Conaway JW / Asturias FJ | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2013 Journal: Nat Struct Mol Biol / Year: 2013Title: A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Authors: Kuang-Lei Tsai / Shigeo Sato / Chieri Tomomori-Sato / Ronald C Conaway / Joan W Conaway / Francisco J Asturias /  Abstract: The CDK8 kinase module (CKM) is a conserved, dissociable Mediator subcomplex whose component subunits were genetically linked to the RNA polymerase II (RNAPII) C-terminal domain (CTD) and ...The CDK8 kinase module (CKM) is a conserved, dissociable Mediator subcomplex whose component subunits were genetically linked to the RNA polymerase II (RNAPII) C-terminal domain (CTD) and individually recognized as transcriptional repressors before Mediator was identified as a pre-eminent complex in eukaryotic transcription regulation. We used macromolecular EM and biochemistry to investigate the subunit organization, structure and Mediator interaction of the Saccharomyces cerevisiae CKM. We found that interaction of the CKM with Mediator's middle module interferes with CTD-dependent RNAPII binding to a previously unknown middle-module CTD-binding site and with the holoenzyme formation process. Taken together, our results reveal the basis for CKM repression, clarify the origin of the connection between CKM subunits and the CTD and suggest that a combination of competitive interactions and conformational changes that facilitate holoenzyme formation underlie the mechanism of transcription regulation by Mediator. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5588.map.gz emd_5588.map.gz | 10.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5588-v30.xml emd-5588-v30.xml emd-5588.xml emd-5588.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5588.jpg emd_5588.jpg | 71.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5588 http://ftp.pdbj.org/pub/emdb/structures/EMD-5588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5588 | HTTPS FTP |
-Validation report
| Summary document |  emd_5588_validation.pdf.gz emd_5588_validation.pdf.gz | 78.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5588_full_validation.pdf.gz emd_5588_full_validation.pdf.gz | 77.3 KB | Display | |
| Data in XML |  emd_5588_validation.xml.gz emd_5588_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5588 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5588 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5588 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5588 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5588.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5588.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of the CDK8 kinase module including Med12, Med13, Cdk8, and CycC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
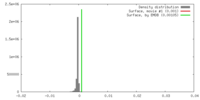
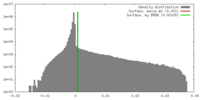
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cdk8 kinase module of yeast Mediator complex
| Entire | Name: Cdk8 kinase module of yeast Mediator complex |
|---|---|
| Components |
|
-Supramolecule #1000: Cdk8 kinase module of yeast Mediator complex
| Supramolecule | Name: Cdk8 kinase module of yeast Mediator complex / type: sample / ID: 1000 / Number unique components: 4 |
|---|---|
| Molecular weight | Experimental: 430 KDa / Theoretical: 430 KDa |
-Macromolecule #1: Med12
| Macromolecule | Name: Med12 / type: protein_or_peptide / ID: 1 / Name.synonym: Srb8 / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 167 KDa / Theoretical: 167 KDa |
-Macromolecule #2: Med13
| Macromolecule | Name: Med13 / type: protein_or_peptide / ID: 2 / Name.synonym: Srb9 / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 160 KDa / Theoretical: 160 KDa |
-Macromolecule #3: Cdk8
| Macromolecule | Name: Cdk8 / type: protein_or_peptide / ID: 3 / Name.synonym: Srb10 / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 63 KDa / Theoretical: 63 KDa |
-Macromolecule #4: CycC
| Macromolecule | Name: CycC / type: protein_or_peptide / ID: 4 / Name.synonym: Srb11 / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 38 KDa / Theoretical: 38 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.020 mg/mL |
|---|---|
| Buffer | pH: 7.6 Details: 20mM HEPES, 200mM potassium acetate, 5mM beta-mercaptoethanol |
| Grid | Details: 300 mesh Cu/Rh grids with continuous carbon, glow discharged in amylamine atmosphere |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 77 K / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 80 K / Max: 110 K / Average: 105 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 125,000 times magnification using a CCD camera |
| Date | Dec 1, 2011 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.35 µm / Number real images: 154 / Average electron dose: 15 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 60400 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.2 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 15.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPARX, SPIDER / Number images used: 70000 |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)