[English] 日本語
 Yorodumi
Yorodumi- EMDB-5581: Negatively Stained TEM structure of trypanosomatid core editing c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5581 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negatively Stained TEM structure of trypanosomatid core editing complex (L-complex) | |||||||||
 Map data Map data | reconstruction of negatively stained L-complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA editing | |||||||||
| Biological species |  Leishmania major (eukaryote) Leishmania major (eukaryote) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Li F / Ge P / Hui WH / Atanasov I / Rogersa K / Guo Q / Osatoa D / Falickd AM / Zhou ZH / Simpson L | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2009 Journal: Proc Natl Acad Sci U S A / Year: 2009Title: Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Authors: Feng Li / Peng Ge / Wong H Hui / Ivo Atanasov / Kestrel Rogers / Qiang Guo / Daren Osato / Arnold M Falick / Z Hong Zhou / Larry Simpson /  Abstract: Uridine insertion/deletion RNA editing is a unique form of posttranscriptional RNA processing that occurs in mitochondria of kinetoplastid protists. We have carried out 3D structural analyses of the ...Uridine insertion/deletion RNA editing is a unique form of posttranscriptional RNA processing that occurs in mitochondria of kinetoplastid protists. We have carried out 3D structural analyses of the core editing complex or "L (ligase)-complex" from Leishmania tarentolae mitochondria isolated by the tandem affinity purification procedure (TAP). The purified material, sedimented at 20-25S, migrated in a blue native gel at 1 MDa and exhibited both precleaved and full-cycle gRNA-mediated U-insertion and U-deletion in vitro activities. The purified L-complex was analyzed by electron tomography to determine the extent of heterogeneity. Three-dimensional structural comparisons of individual particles in the tomograms revealed that a majority of the complexes have a similar shape of a slender triangle. An independent single-particle reconstruction, using a featureless Gaussian ball as the initial model, converged to a similar triangular structure. Another single-particle reconstruction, using the averaged tomography structure as the initial model, yielded a similar structure. The REL1 ligase was localized on the model to the base of the apex by decoration with REL1-specific IgG. This structure should prove useful for a detailed analysis of the editing reaction. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5581.map.gz emd_5581.map.gz | 20.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5581-v30.xml emd-5581-v30.xml emd-5581.xml emd-5581.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5581_1.jpg emd_5581_1.jpg | 58.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5581 http://ftp.pdbj.org/pub/emdb/structures/EMD-5581 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5581 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5581 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5581.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5581.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | reconstruction of negatively stained L-complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
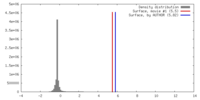
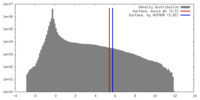
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : L-Complex from L. major
| Entire | Name: L-Complex from L. major |
|---|---|
| Components |
|
-Supramolecule #1000: L-Complex from L. major
| Supramolecule | Name: L-Complex from L. major / type: sample / ID: 1000 / Oligomeric state: A single complex / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 1.0 MDa / Method: blue native gel |
-Macromolecule #1: Core editing complex
| Macromolecule | Name: Core editing complex / type: protein_or_peptide / ID: 1 / Name.synonym: L-Complex / Number of copies: 1 / Oligomeric state: complex of many subunits / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Leishmania major (eukaryote) / synonym: Leishmania / Organelle: Mitochondria Leishmania major (eukaryote) / synonym: Leishmania / Organelle: Mitochondria |
| Molecular weight | Experimental: 1.0 MDa |
| Recombinant expression | Organism:  Leishmania tarentolae (eukaryote) / Recombinant plasmid: pMRP1-TAP Leishmania tarentolae (eukaryote) / Recombinant plasmid: pMRP1-TAP |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 / Details: 20 mM Tris, pH 7.6, 60 mM KCl, 10 mM MgCl2 |
|---|---|
| Staining | Type: NEGATIVE / Details: 2% uranyl acetate for 2 min |
| Grid | Details: continuous carbon grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 300 K |
| Date | Aug 1, 2008 |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD / Digitization - Sampling interval: 15 µm / Number real images: 26 / Average electron dose: 400 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 68200 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.6 µm / Nominal magnification: 70000 |
| Sample stage | Specimen holder: Single tilt room temperature holder / Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: phase flip |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 12500 |
| Final two d classification | Number classes: 879 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)