[English] 日本語
 Yorodumi
Yorodumi- EMDB-5368: Structure of endophilin bound to a membrane tubule with a diamete... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5368 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
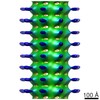
| Title | Structure of endophilin bound to a membrane tubule with a diameter of 32nm | |||||||||
 Map data Map data | Structure of Endophilin bound to the bilayer | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 26.0 Å | |||||||||
 Authors Authors | Mim C / Cui H / Gawronski-Salerno JA / Frost A / Lyman E / Voth GA / Unger VM | |||||||||
 Citation Citation |  Journal: Cell / Year: 2012 Journal: Cell / Year: 2012Title: Structural basis of membrane bending by the N-BAR protein endophilin. Authors: Carsten Mim / Haosheng Cui / Joseph A Gawronski-Salerno / Adam Frost / Edward Lyman / Gregory A Voth / Vinzenz M Unger /  Abstract: Functioning as key players in cellular regulation of membrane curvature, BAR domain proteins bend bilayers and recruit interaction partners through poorly understood mechanisms. Using electron ...Functioning as key players in cellular regulation of membrane curvature, BAR domain proteins bend bilayers and recruit interaction partners through poorly understood mechanisms. Using electron cryomicroscopy, we present reconstructions of full-length endophilin and its N-terminal N-BAR domain in their membrane-bound state. Endophilin lattices expose large areas of membrane surface and are held together by promiscuous interactions between endophilin's amphipathic N-terminal helices. Coarse-grained molecular dynamics simulations reveal that endophilin lattices are highly dynamic and that the N-terminal helices are required for formation of a stable and regular scaffold. Furthermore, endophilin accommodates different curvatures through a quantized addition or removal of endophilin dimers, which in some cases causes dimerization of endophilin's SH3 domains, suggesting that the spatial presentation of SH3 domains, rather than affinity, governs the recruitment of downstream interaction partners. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5368.map.gz emd_5368.map.gz | 12.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5368-v30.xml emd-5368-v30.xml emd-5368.xml emd-5368.xml | 9.3 KB 9.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5368_1.jpg emd_5368_1.jpg | 61.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5368 http://ftp.pdbj.org/pub/emdb/structures/EMD-5368 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5368 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5368 | HTTPS FTP |
-Validation report
| Summary document |  emd_5368_validation.pdf.gz emd_5368_validation.pdf.gz | 79 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5368_full_validation.pdf.gz emd_5368_full_validation.pdf.gz | 78.1 KB | Display | |
| Data in XML |  emd_5368_validation.xml.gz emd_5368_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5368 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5368 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5368 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5368 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5368.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5368.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Endophilin bound to the bilayer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.73 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Structure of Endophilin bound to the bilayer
| Entire | Name: Structure of Endophilin bound to the bilayer |
|---|---|
| Components |
|
-Supramolecule #1000: Structure of Endophilin bound to the bilayer
| Supramolecule | Name: Structure of Endophilin bound to the bilayer / type: sample / ID: 1000 / Oligomeric state: Dimer of endophilin bound to the bilayer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 37 MDa / Method: SDS-PAGE |
-Macromolecule #1: endophilin-A1
| Macromolecule | Name: endophilin-A1 / type: protein_or_peptide / ID: 1 / Name.synonym: endophilin / Number of copies: 2 / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 37 MDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 50mM potassium aspartate, 10mM Tris/HCl, 1mM EGTA |
| Grid | Details: C-Flat, 2-2, 400nm mesh |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 105 K / Instrument: OTHER / Details: Vitrification instrument: FEI Vitrobot |
| Details | protein and liposomes added in solution before freezing |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | Jun 17, 2009 |
| Image recording | Average electron dose: 10 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder: eucentric, single tilt / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 17.27 Å Applied symmetry - Helical parameters - Δ&Phi: 59.18 ° Applied symmetry - Helical parameters - Axial symmetry: C2 (2 fold cyclic) Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 26.0 Å / Resolution method: OTHER / Software - Name: SPIDER,XMIPP,IHRSR |
|---|---|
| CTF correction | Details: ACE,MATLAB |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)