+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5353 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The 3D volume of the Birnavirus, Espirito Santo Virus. | |||||||||
 Map data Map data | This is the 3D volume of the Birnavirus, Espirito Santo Virus | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Birnavirus / cryoEM | |||||||||
| Biological species |  Espirito Santo virus Espirito Santo virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 12.0 Å | |||||||||
 Authors Authors | Vancini R / Paredes A / Ribeiro M / Blackburn K / Ferreira D / Kononchik J / Hernandez R / Brown D | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2012 Journal: J Virol / Year: 2012Title: Espirito Santo virus: a new birnavirus that replicates in insect cells. Authors: Ricardo Vancini / Angel Paredes / Mariana Ribeiro / Kevin Blackburn / Davis Ferreira / Joseph P Kononchik / Raquel Hernandez / Dennis Brown /  Abstract: Espirito Santo virus (ESV) is a newly discovered virus recovered as contamination in a sample of a virulent strain of dengue-2 virus (strain 44/2), which was recovered from a patient in the state of ...Espirito Santo virus (ESV) is a newly discovered virus recovered as contamination in a sample of a virulent strain of dengue-2 virus (strain 44/2), which was recovered from a patient in the state of Espirito Santo, Brazil, and amplified in insect cells. ESV was found to be dependent upon coinfection with a virulent strain of dengue-2 virus and to replicate in C6/36 insect cells but not in mammalian Vero cells. A sequence of the genome has been produced by de novo assembly and was not found to match to any known viral sequence. An incomplete match to the nucleotide sequence of the RNA-dependent RNA polymerase from Drosophila X virus (DXV), another birnavirus, could be detected. Mass spectrometry analysis of ESV proteins found no matches in the protein data banks. However, peptides recovered by mass spectrometry corresponded to the de novo-assembled sequence by BLAST analysis. The composition and three-dimensional structure of ESV are presented, and its sequence is compared to those of other members of the birnavirus family. Although the virus was found to belong to the family Birnaviridae, biochemical and sequence information for ESV differed from that of DXV, the representative species of the genus Entomobirnavirus. Thus, significant differences underscore the uniqueness of this infectious agent, and its relationship to the coinfecting virus is discussed. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5353.map.gz emd_5353.map.gz | 54.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5353-v30.xml emd-5353-v30.xml emd-5353.xml emd-5353.xml | 9.2 KB 9.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5353_1.jpg emd_5353_1.jpg | 84.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5353 http://ftp.pdbj.org/pub/emdb/structures/EMD-5353 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5353 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5353 | HTTPS FTP |
-Validation report
| Summary document |  emd_5353_validation.pdf.gz emd_5353_validation.pdf.gz | 78.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5353_full_validation.pdf.gz emd_5353_full_validation.pdf.gz | 77.2 KB | Display | |
| Data in XML |  emd_5353_validation.xml.gz emd_5353_validation.xml.gz | 492 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5353 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5353 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5353 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5353 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5353.map.gz / Format: CCP4 / Size: 141.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5353.map.gz / Format: CCP4 / Size: 141.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the 3D volume of the Birnavirus, Espirito Santo Virus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.29 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
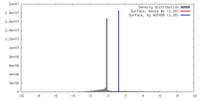
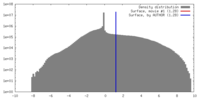
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Espirito Santo Virus
| Entire | Name:  Espirito Santo Virus Espirito Santo Virus |
|---|---|
| Components |
|
-Supramolecule #1000: Espirito Santo Virus
| Supramolecule | Name: Espirito Santo Virus / type: sample / ID: 1000 / Oligomeric state: Icosahedral / Number unique components: 1 |
|---|
-Supramolecule #1: Espirito Santo virus
| Supramolecule | Name: Espirito Santo virus / type: virus / ID: 1 / Name.synonym: ESV / NCBI-ID: 1127767 / Sci species name: Espirito Santo virus / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No / Syn species name: ESV |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: INVERTEBRATES Homo sapiens (human) / synonym: INVERTEBRATES |
| Virus shell | Shell ID: 1 / Diameter: 750 Å / T number (triangulation number): 13 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 1.5% glutaraldehyde, 20mM HEPES, pH 7.5 |
|---|---|
| Grid | Details: 400 mesh copper grids |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: Home made plunger. Vitrification carried out using home made lacey carbon films according to previously published protocols Method: Applied specimen to lacey carbon grids, blotted from behind and plunged into cryogen after 2-3 seconds |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: corrected using power spectrum |
| Date | Jun 20, 2010 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Digitization - Sampling interval: 15 µm / Number real images: 199 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 39000 |
| Sample stage | Specimen holder: Screw ring cartidge / Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were selected using e2boxer.py in the EMAN2 suite of programs |
|---|---|
| CTF correction | Details: Each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 12.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 5928 |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)