+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6969 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Aquareovirus | ||||||||||||||||||
 Map data Map data | Icosahedral capsid of aquareovirus | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | icosahedral capsid / symmetry-mismatch / genome / RNA-dependent RNA polymerase / VIRUS | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont entry into host cell via permeabilization of inner membrane / permeabilization of host organelle membrane involved in viral entry into host cell / host cell surface binding / viral inner capsid / viral outer capsid / 7-methylguanosine mRNA capping / viral capsid / mRNA guanylyltransferase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity ...symbiont entry into host cell via permeabilization of inner membrane / permeabilization of host organelle membrane involved in viral entry into host cell / host cell surface binding / viral inner capsid / viral outer capsid / 7-methylguanosine mRNA capping / viral capsid / mRNA guanylyltransferase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / hydrolase activity / RNA helicase / GTP binding / ATP binding / metal ion binding Similarity search - Function | ||||||||||||||||||
| Biological species |  Grass carp reovirus Grass carp reovirus | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||||||||
 Authors Authors | Liu H / Fang Q | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly. Authors: Xurong Wang / Fuxian Zhang / Rui Su / Xiaowu Li / Wenyuan Chen / Qingxiu Chen / Tao Yang / Jiawei Wang / Hongrong Liu / Qin Fang / Lingpeng Cheng /  Abstract: Most double-stranded RNA (dsRNA) viruses transcribe RNA plus strands within a common innermost capsid shell. This process requires coordinated efforts by RNA-dependent RNA polymerase (RdRp) together ...Most double-stranded RNA (dsRNA) viruses transcribe RNA plus strands within a common innermost capsid shell. This process requires coordinated efforts by RNA-dependent RNA polymerase (RdRp) together with other capsid proteins and genomic RNA. Here we report the near-atomic resolution structure of the RdRp protein VP2 in complex with its cofactor protein VP4 and genomic RNA within an aquareovirus capsid using 200-kV cryoelectron microscopy and symmetry-mismatch reconstruction. The structure of these capsid proteins enabled us to observe the elaborate nonicosahedral structure within the double-layered icosahedral capsid. Our structure shows that the RdRp complex is anchored at the inner surface of the capsid shell and interacts with genomic dsRNA and four of the five asymmetrically arranged N termini of the capsid shell proteins under the fivefold axis, implying roles for these N termini in virus assembly. The binding site of the RNA end at VP2 is different from the RNA cap binding site identified in the crystal structure of orthoreovirus RdRp λ3, although the structures of VP2 and λ3 are almost identical. A loop, which was thought to separate the RNA template and transcript, interacts with an apical domain of the capsid shell protein, suggesting a mechanism for regulating RdRp replication and transcription. A conserved nucleoside triphosphate binding site was localized in our RdRp cofactor protein VP4 structure, and interactions between the VP4 and the genomic RNA were identified. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6969.map.gz emd_6969.map.gz | 2.5 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6969-v30.xml emd-6969-v30.xml emd-6969.xml emd-6969.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6969.png emd_6969.png | 96.6 KB | ||
| Masks |  emd_6969_msk_1.map emd_6969_msk_1.map | 11.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-6969.cif.gz emd-6969.cif.gz | 8.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6969 http://ftp.pdbj.org/pub/emdb/structures/EMD-6969 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6969 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6969 | HTTPS FTP |
-Validation report
| Summary document |  emd_6969_validation.pdf.gz emd_6969_validation.pdf.gz | 795.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6969_full_validation.pdf.gz emd_6969_full_validation.pdf.gz | 795.3 KB | Display | |
| Data in XML |  emd_6969_validation.xml.gz emd_6969_validation.xml.gz | 11.2 KB | Display | |
| Data in CIF |  emd_6969_validation.cif.gz emd_6969_validation.cif.gz | 13.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6969 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6969 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6969 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6969 | HTTPS FTP |
-Related structure data
| Related structure data |  5zvtMC  6968C  5zvsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6969.map.gz / Format: CCP4 / Size: 2.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6969.map.gz / Format: CCP4 / Size: 2.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Icosahedral capsid of aquareovirus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
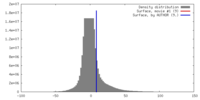
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_6969_msk_1.map emd_6969_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Grass carp reovirus
| Entire | Name:  Grass carp reovirus Grass carp reovirus |
|---|---|
| Components |
|
-Supramolecule #1: Grass carp reovirus
| Supramolecule | Name: Grass carp reovirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 / NCBI-ID: 128987 / Sci species name: Grass carp reovirus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Outer capsid VP7
| Macromolecule | Name: Outer capsid VP7 / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Grass carp reovirus Grass carp reovirus |
| Molecular weight | Theoretical: 29.844648 KDa |
| Recombinant expression | Organism:  Ctenopharyngodon idella (grass carp) Ctenopharyngodon idella (grass carp) |
| Sequence | String: MPLHMIPQVA HAMVRAAAAG RLTLYTRTRT ETTNFDHAEY VTCGRYTICA FCLTTLAPHA NVKTIQDSHA CSRQPNEAIR SLVEVSDKA QTALVGSRTV DYHELDVKAG FVAPTADETI APSKDIVELP FRTCDLDDSS ATACVRNHCQ AGHDGVIHLP I LSGDFKLP ...String: MPLHMIPQVA HAMVRAAAAG RLTLYTRTRT ETTNFDHAEY VTCGRYTICA FCLTTLAPHA NVKTIQDSHA CSRQPNEAIR SLVEVSDKA QTALVGSRTV DYHELDVKAG FVAPTADETI APSKDIVELP FRTCDLDDSS ATACVRNHCQ AGHDGVIHLP I LSGDFKLP NEHPTKPLDD THPHDKVLTR CPKTGLLLVH DTHAHATAVV ATAATRAILM HDLLTSANAD DGHQARSACY GP AFNNLTF ACHSTCASDM AHFDCGQIVG LDLHVEPSD UniProtKB: Outer capsid VP7 |
-Macromolecule #2: N-terminus of outer capsid protein VP5
| Macromolecule | Name: N-terminus of outer capsid protein VP5 / type: protein_or_peptide / ID: 2 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Grass carp reovirus Grass carp reovirus |
| Molecular weight | Theoretical: 4.302686 KDa |
| Recombinant expression | Organism:  Ctenopharyngodon idella (grass carp) Ctenopharyngodon idella (grass carp) |
| Sequence | String: MGNVQTSVNT YNITGDGNSF TPTSDMTSTA APAIDLKPGV LN UniProtKB: Putative outer capsid VP4 |
-Macromolecule #3: C-terminus of outer capsid protein VP5
| Macromolecule | Name: C-terminus of outer capsid protein VP5 / type: protein_or_peptide / ID: 3 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Grass carp reovirus Grass carp reovirus |
| Molecular weight | Theoretical: 64.362086 KDa |
| Recombinant expression | Organism:  Ctenopharyngodon idella (grass carp) Ctenopharyngodon idella (grass carp) |
| Sequence | String: PTGKLWRPVG TSVATIDSLA IVSDRFGQYS FVNEGMRETF SKALFDINMW QPLFQATKTG CGPIVLSSFT TTTSGYVGAT AGDALDNPV TNGVFISTVQ IMNLQRTIAA RMRDVALWQK HLDTAMTMLT PDISAGSASC NWKSLLAFAK DILPLDNLCL T YPNEFYNV ...String: PTGKLWRPVG TSVATIDSLA IVSDRFGQYS FVNEGMRETF SKALFDINMW QPLFQATKTG CGPIVLSSFT TTTSGYVGAT AGDALDNPV TNGVFISTVQ IMNLQRTIAA RMRDVALWQK HLDTAMTMLT PDISAGSASC NWKSLLAFAK DILPLDNLCL T YPNEFYNV AIHRYPALKP GNPDTKLPDA QAHPLGEVAG AFNAATSEVG SLVGSSSTLS QAISTMAGKD LDLIEADTPL PV SVFTPSL APRSYRPAFI KPEDAKWIAE FNNSSLIRKT LTYSGATYTV QLGPGPTRVI DMNAMIDSVL TLDVSGTILP YDT NPDLST SVPAFVLIQT SVPIQQVTTA ANITAITVVS AAGASAINLA INVRGQPRFN MLHLQATFER ETITGIPYIY GLGT FLIPS PTSSSNFSNP TLMDGLLTVT PVLLRETTYK GEVVDAIVPA TVMANQTSEE VASALANDAI VLVSNHLNKL ANVVG DAIP VASRTDDSAT SAIVSRLAVQ HKLSQVGQAS PTPPDYPLLW RRAKRAASMF VSNPSLALQV GIPVLTQSGM LSALTS GVG TALRTGSLGK GVTDASEKLR ARQSLTVAKQ AFFDQIGSLW PGK UniProtKB: Putative outer capsid VP4 |
-Macromolecule #4: Core protein VP6
| Macromolecule | Name: Core protein VP6 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Grass carp reovirus Grass carp reovirus |
| Molecular weight | Theoretical: 44.606535 KDa |
| Recombinant expression | Organism:  Ctenopharyngodon idella (grass carp) Ctenopharyngodon idella (grass carp) |
| Sequence | String: MAQRQFFGLT YNFYGQPAPL FDLNDLQELA GCYARPWTSR FSHLAISTGS LPVWSARYPS VASRNIVVNT LLGAHLNPFA GGQITSHQG ITWRDPVLSS LAPVPAIQPP PVWAVAENVL LDSNNYPTYV LNLSSMWPIN QDVHIMTMWA LSDQGPIYHL E VPVDPMPA ...String: MAQRQFFGLT YNFYGQPAPL FDLNDLQELA GCYARPWTSR FSHLAISTGS LPVWSARYPS VASRNIVVNT LLGAHLNPFA GGQITSHQG ITWRDPVLSS LAPVPAIQPP PVWAVAENVL LDSNNYPTYV LNLSSMWPIN QDVHIMTMWA LSDQGPIYHL E VPVDPMPA ATTAALMAYT GVPIAHLAQT AYRFAGQLPQ SPDSTMVSTI RWLSAIWFGS LTGRLNRSRT CNGFYFEFAK PA LNPDQAV LKWNDGARAA PPAAAQSSYI RCISPHWQHQ IVEVAGALMS QSVTAVTGLP ALIDEATLPA WSQGVANLTG NGQ GVVPCL DYNPVPMAAA RHLQWRQDGL ITAAQEAQLN NDYTAYALTI ERHLTAMLVA NPIAAGRMPI QPFNAADFGQ AGQT AAAVA LAQAMFV UniProtKB: Core protein VP6 |
-Macromolecule #5: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Grass carp reovirus Grass carp reovirus |
| Molecular weight | Theoretical: 141.512156 KDa |
| Recombinant expression | Organism:  Ctenopharyngodon idella (grass carp) Ctenopharyngodon idella (grass carp) |
| Sequence | String: MAAVFGIQLV PKLNTSTTRR TFLPLRFDLL LDRLQSTNLH GVLYRALDFN PVDRSATVIQ TYPPLNAWSP HHAFIENPLD YRDWTEFIH DRALAFVGVL TQRYPLTQNA QRYTNPLVLG AAFGDFLNAR SIDIFLDRLF YDPTQDSPIT AITKFPYQWT I DSNVTTDS ...String: MAAVFGIQLV PKLNTSTTRR TFLPLRFDLL LDRLQSTNLH GVLYRALDFN PVDRSATVIQ TYPPLNAWSP HHAFIENPLD YRDWTEFIH DRALAFVGVL TQRYPLTQNA QRYTNPLVLG AAFGDFLNAR SIDIFLDRLF YDPTQDSPIT AITKFPYQWT I DSNVTTDS VRTSAGCKYI TLYGYDPSRP STPATYGKHR PTYATVFYYS TLPARSRLLA NLAAGPTVLE HFDSPTYGPH LL LPQTGDV LGYSSSLISQ AALLMVESVM DALRDNANAS ASTAVTRLDQ SYHPVTSFDP STFNTLLQRA TNLALLAVQG VQS ESAIPA IPTMSDVRSF VARLMAEGDP QQWFPYRVDQ ILYWPESPFV PPIGPFYAPF RPVNFPFTTG SYTVVPDASR PLRL LPQYR NATITVQQAD DAYEDTALSP LITTHGFCVT GGVFTSIYDI SGDPTAYPPA QLVDAPNDYF DRERMARRDL FRRLR APAD RSAIKDRAVF DFLASLVNPT TANPVLDTSF SMAYLGASSA HANADEPVIL ADIRSGSIPG LPIPRRIVQF GYDVVH GSL LDLSRAVPTG TFGLVYADLD QVEDAGTDMP AANRAAIAML GTALQMTTAG GVSVLKVNFP TRAFWTQVFN LYATHAT TL HLVKPTIVNS SEVFLVFGGR QSNGALRSTT ALQRALLSLY ARNAAIDRAV THIPFFGVPD DGTSDLGIDA VRLFDPMF S DAVANLPSNA LASLVSRVVP SSIMFTRVPS NGPVSTTIYG KRTFLSNRRR ARLRDVPMLI TTTLVHQRRF TTPPTFTLF SSEAVPVTTL VAAGYNSFIS EQTRNPNLAH LLDLGTGPEC RILSLIPPTL QVTMSDSRPC AELMASFDPA LTAYVQGDYS TAAFWNGIR CDSATAIFTI GAAAAAAGTD LIAFVQQLIP RIVAAGGTRM WLQLNTPLYE VSSLPDLIEI DLRDHVYRFN G GERVEPYA DPVPLQQAIA ALLPAAALSW HTLSPTCDWL PYIIGVGSPL NLSDINTAIS YSRLTPILHI DTTTPPLRVN PV PTPLNQQ CAIRITSLDP AAVLSVQHNG VEVIGGTPGN VISVAGAAAL QYILANQEFL LQFTPTLPGI FDVFLTTLGQ PPV PRGSFT ITPPPTTVAL NMPPPRQLDF TDVGNDARIT CDPYYQLAVC IFKDGQYVRV NPEKASVVTN APNRDLHFVL DLAD NHVLL YLCDVTPSGL GDRIAFPIVD IYRIAFPRNT PVRASLPYTG GGAHLTSGGN PFMSLTTPPA VLPAGVALAA LSTSV ATQY PTYTLPAGVY EYVIE UniProtKB: VP1 |
-Macromolecule #6: VP3
| Macromolecule | Name: VP3 / type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Grass carp reovirus Grass carp reovirus |
| Molecular weight | Theoretical: 132.203312 KDa |
| Recombinant expression | Organism:  Ctenopharyngodon idella (grass carp) Ctenopharyngodon idella (grass carp) |
| Sequence | String: MPRRSARKAQ SAIASPADTN VVPAKDAPTT NSPPSTTSPN QAAADANQQQ AGIVSSQSGP NAVGDSAPSS SVNNDGDIIT RPTSDSIAA VANATKPAAV VSDPQSMKVT PIVNPSSYVC NVCNARFSTM SALSEHLRSD HRDDASTLLA TPMINNAIRS F LTAWDDIR ...String: MPRRSARKAQ SAIASPADTN VVPAKDAPTT NSPPSTTSPN QAAADANQQQ AGIVSSQSGP NAVGDSAPSS SVNNDGDIIT RPTSDSIAA VANATKPAAV VSDPQSMKVT PIVNPSSYVC NVCNARFSTM SALSEHLRSD HRDDASTLLA TPMINNAIRS F LTAWDDIR ILSPDVSSKS LSAYLDSAVA NGPELIIEDT GLCTSFMLLD NIPSAHLTKE LIGFTWFMQM YQMTPPLPEG AV NRIVCMT NWASLGDEGR GLEVRLPPPT DSSVHAYKTV LSRGYIDNAQ FNPLALRSNV LLMLLQFTLS NLKINKSSTF TSD VTTITS GRMIRAFEGR PELLALAYPG RAVLPTQTKN AQFLSTAIAD RIGRLDRANL IGGEVSAMVE CMELCDALTL HIRE TYIML LRSMHQDPTQ IVQIVNECAN NLLNSTIPIS LRPTILCPWF ASSEDLRLQQ VMHLVNISSN TAAALPLVEA LSTLL RSVT PLVLDPTVLT NAITTISEST TQTISPISEI LRLLQPMGND YAAFWKCIAS WAYNGLVTTV LSEDAFPDSS QSITHL PSM WKCLFLTLAG PMTSDPHSPV KVFMALANLL AQPEPIAIGV PGMHQTTPAS QFSHPGVWPP GFLNPQLINP QQAPLLR AF AEHIRANWPQ PSEFGYGSTL QGSANLFIPS NRMVYPWPNQ PLPRLTVAPT YDSAMSNWIS TTIAFFIRVV NSVNMTAT V NDLTRRTMTG VMTAMRQVKT MTPFYIQHMC PTELSVLASV TVTPPFQVPF TRLVQNDVIT NVLVARVDPA QRGDAAVDI RATHATFAAA LPVDPAAIVV AMLCGQTETN LIPSHHYGKA FAPLFASNAM FTRNQRAVIT REAFVCARSA VAQCQDAGFL VPRPLDALR QFDVTSAAAA EIMHAVNDAF KTAFDLDGAL LDGLALYGDP RIADLSAAYL QYGGNVVREH VPPGPSHIHR A LQQVESTF MAEMNLFNVA RGNLYLVQTA TNGNWSPMAP VAAPPFVRGG PNVRVVGRFG TIVPRPNGLE PQLIDDGNVP RD IAGDWVY PSDVLQVSVA VFRDYVWPMV KAGRTRVLVE LGHYVYTLHY YDPQISLDEA PILEEWLSKI NPAGIPPVPF CIP IPQVYP CITARRVHYA FTSENNNDSL FSTNAASIDT AFGENAAVSP LRWPGLVDPN YRVGTNDLPN RITLYNSLYR YNFT YPTLD GIMYVRSAT UniProtKB: RNA helicase |
-Macromolecule #7: MYRISTIC ACID
| Macromolecule | Name: MYRISTIC ACID / type: ligand / ID: 7 / Number of copies: 10 / Formula: MYR |
|---|---|
| Molecular weight | Theoretical: 228.371 Da |
| Chemical component information |  ChemComp-MYR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)