+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5342 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
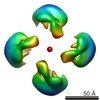
| Title | DNA-free p53 Tetramer Reconstruction | |||||||||
 Map data Map data | DNA-free p53 Tetramer Reconstruction | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining | |||||||||
 Authors Authors | Pham N / Lucumi A / Cheung N / Viadiu H | |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2012 Journal: Biochemistry / Year: 2012Title: The tetramer of p53 in the absence of DNA forms a relaxed quaternary state. Authors: Nam Pham / Armando Lucumi / Nikki Cheung / Hector Viadiu /  Abstract: p53 is a tetrameric multidomain protein that triggers the anticancer cellular response to stress. We have calculated a three-dimensional reconstruction of full-length human p53 in the absence of DNA ...p53 is a tetrameric multidomain protein that triggers the anticancer cellular response to stress. We have calculated a three-dimensional reconstruction of full-length human p53 in the absence of DNA using single-particle electron microscopy. The reconstruction of DNA-free full-length p53 shows a square-shaped structure with four distinct domains and a hollow center. In comparison with the known compacted DNA-bound full-length p53 structures, the DNA-free p53 tetramer adopts a relaxed conformation with separated monomers and oligomerization interfaces different from those of the DNA-bound conformation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5342.map.gz emd_5342.map.gz | 5.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5342-v30.xml emd-5342-v30.xml emd-5342.xml emd-5342.xml | 7.4 KB 7.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5342_1.jpg emd_5342_1.jpg | 69.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5342 http://ftp.pdbj.org/pub/emdb/structures/EMD-5342 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5342 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5342 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5342.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5342.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DNA-free p53 Tetramer Reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : p53
| Entire | Name: p53 |
|---|---|
| Components |
|
-Supramolecule #1000: p53
| Supramolecule | Name: p53 / type: sample / ID: 1000 / Oligomeric state: tetramer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 200 KDa / Theoretical: 200 KDa / Method: Gel filtration |
-Macromolecule #1: p53
| Macromolecule | Name: p53 / type: protein_or_peptide / ID: 1 / Name.synonym: p53 / Number of copies: 4 / Oligomeric state: Tetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Location in cell: Nucleus Homo sapiens (human) / synonym: Human / Location in cell: Nucleus |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: 150mM NaCl |
| Staining | Type: NEGATIVE / Details: 1% uranyl formate |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPHERA |
|---|---|
| Image recording | Number real images: 50 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: PHILIPS ROTATION HOLDER |
- Image processing
Image processing
| Final reconstruction | Number images used: 8550 |
|---|---|
| Final two d classification | Number classes: 50 |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera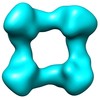





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















