+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6012 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron microscopy of Calsyntenin-3 tetramer conformation II | |||||||||
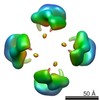
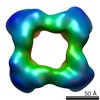
 Map data Map data | Cstn-3 High Molecular Weight (HMW) tetramer conformation II | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Synaptic organizer / trans-synaptic bridge | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 16.5 Å | |||||||||
 Authors Authors | Lu Z / Wang Y / Chen F / Tong F / Reddy MVVVS / Luo L / Seshadrinathan S / Zhang L / Holthauzen LMF / Craig AM ...Lu Z / Wang Y / Chen F / Tong F / Reddy MVVVS / Luo L / Seshadrinathan S / Zhang L / Holthauzen LMF / Craig AM / Ren G / Rudenko G | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2014 Journal: J Biol Chem / Year: 2014Title: Calsyntenin-3 molecular architecture and interaction with neurexin 1α. Authors: Zhuoyang Lu / Yun Wang / Fang Chen / Huimin Tong / M V V V Sekhar Reddy / Lin Luo / Suchithra Seshadrinathan / Lei Zhang / Luis Marcelo F Holthauzen / Ann Marie Craig / Gang Ren / Gabby Rudenko /    Abstract: Calsyntenin 3 (Cstn3 or Clstn3), a recently identified synaptic organizer, promotes the development of synapses. Cstn3 localizes to the postsynaptic membrane and triggers presynaptic differentiation. ...Calsyntenin 3 (Cstn3 or Clstn3), a recently identified synaptic organizer, promotes the development of synapses. Cstn3 localizes to the postsynaptic membrane and triggers presynaptic differentiation. Calsyntenin members play an evolutionarily conserved role in memory and learning. Cstn3 was recently shown in cell-based assays to interact with neurexin 1α (n1α), a synaptic organizer that is implicated in neuropsychiatric disease. Interaction would permit Cstn3 and n1α to form a trans-synaptic complex and promote synaptic differentiation. However, it is contentious whether Cstn3 binds n1α directly. To understand the structure and function of Cstn3, we determined its architecture by electron microscopy and delineated the interaction between Cstn3 and n1α biochemically and biophysically. We show that Cstn3 ectodomains form monomers as well as tetramers that are stabilized by disulfide bonds and Ca(2+), and both are probably flexible in solution. We show further that the extracellular domains of Cstn3 and n1α interact directly and that both Cstn3 monomers and tetramers bind n1α with nanomolar affinity. The interaction is promoted by Ca(2+) and requires minimally the LNS domain of Cstn3. Furthermore, Cstn3 uses a fundamentally different mechanism to bind n1α compared with other neurexin partners, such as the synaptic organizer neuroligin 2, because Cstn3 does not strictly require the sixth LNS domain of n1α. Our structural data suggest how Cstn3 as a synaptic organizer on the postsynaptic membrane, particularly in tetrameric form, may assemble radially symmetric trans-synaptic bridges with the presynaptic synaptic organizer n1α to recruit and spatially organize proteins into networks essential for synaptic function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6012.map.gz emd_6012.map.gz | 5.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6012-v30.xml emd-6012-v30.xml emd-6012.xml emd-6012.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6012.gif 400_6012.gif 80_6012.gif 80_6012.gif | 15.4 KB 2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6012 http://ftp.pdbj.org/pub/emdb/structures/EMD-6012 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6012 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6012 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6012.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6012.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cstn-3 High Molecular Weight (HMW) tetramer conformation II | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.88 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
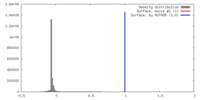
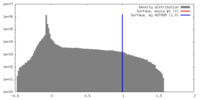
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cstn-3 Tetramer (High Molecular Weight)
| Entire | Name: Cstn-3 Tetramer (High Molecular Weight) |
|---|---|
| Components |
|
-Supramolecule #1000: Cstn-3 Tetramer (High Molecular Weight)
| Supramolecule | Name: Cstn-3 Tetramer (High Molecular Weight) / type: sample / ID: 1000 / Oligomeric state: Tetramer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 352 KDa Method: Calculated from amino acid content without N-linked glycosylation |
-Macromolecule #1: Calsyntenin-3
| Macromolecule | Name: Calsyntenin-3 / type: protein_or_peptide / ID: 1 / Name.synonym: Cstn-3 / Number of copies: 4 / Oligomeric state: Tetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Tissue: neuron / Location in cell: Synapse Homo sapiens (human) / synonym: Human / Tissue: neuron / Location in cell: Synapse |
| Molecular weight | Theoretical: 88 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) / Recombinant strain: BTI-TN-5B1-4 / Recombinant cell: High Five Trichoplusia ni (cabbage looper) / Recombinant strain: BTI-TN-5B1-4 / Recombinant cell: High Five |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 25 mM Tris, pH 8.0, 120 mM NaCl, 5 mM CaCl2 |
| Staining | Type: NEGATIVE Details: An aliquot (~4 uL) of sample was placed on a grid. After ~1 min incubation, excess solution was blotted with filter paper, and the grid stained for ~1 min by submersion in one drop (~35 uL) ...Details: An aliquot (~4 uL) of sample was placed on a grid. After ~1 min incubation, excess solution was blotted with filter paper, and the grid stained for ~1 min by submersion in one drop (~35 uL) of 1% w/v uranyl formate (UF) on Parafilm before being nitrogen-air-dried at room temperature. |
| Grid | Details: thin-carbon-coated 200 mesh copper grid (CF200-Cu, EMS), glow-discharged 15 seconds |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | ZEISS LIBRA120PLUS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 125,000 times magnification. |
| Date | Dec 11, 2012 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 639 / Average electron dose: 200 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 0.6 µm / Nominal defocus min: 0.1 µm / Nominal magnification: 125000 |
| Sample stage | Specimen holder model: OTHER |
- Image processing
Image processing
| CTF correction | Details: ctffind3 |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 16.5 Å / Resolution method: OTHER / Software - Name: EMAN, IPET / Number images used: 6699 |
| Final two d classification | Number classes: 150 |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)