[English] 日本語
 Yorodumi
Yorodumi- EMDB-5314: Structures of the RNA-guided surveillance complex from a bacteria... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5314 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structures of the RNA-guided surveillance complex from a bacterial immune system | |||||||||
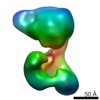
 Map data Map data | reconstructed density of the E. coli cascade complex at 8 Angstroms resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cascade / bacterial immune system / CRISPR / RNA-guided / ribo-nucleoprotein / ssRNA | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.8 Å | |||||||||
 Authors Authors | Wiedenheft B / Lander GC / Zhou K / Jore MM / Brouns SJJ / van der Oost J / Doudna JA / Nogales E | |||||||||
 Citation Citation |  Journal: Nature / Year: 2011 Journal: Nature / Year: 2011Title: Structures of the RNA-guided surveillance complex from a bacterial immune system. Authors: Blake Wiedenheft / Gabriel C Lander / Kaihong Zhou / Matthijs M Jore / Stan J J Brouns / John van der Oost / Jennifer A Doudna / Eva Nogales /   Abstract: Bacteria and archaea acquire resistance to viruses and plasmids by integrating short fragments of foreign DNA into clustered regularly interspaced short palindromic repeats (CRISPRs). These ...Bacteria and archaea acquire resistance to viruses and plasmids by integrating short fragments of foreign DNA into clustered regularly interspaced short palindromic repeats (CRISPRs). These repetitive loci maintain a genetic record of all prior encounters with foreign transgressors. CRISPRs are transcribed and the long primary transcript is processed into a library of short CRISPR-derived RNAs (crRNAs) that contain a unique sequence complementary to a foreign nucleic-acid challenger. In Escherichia coli, crRNAs are incorporated into a multisubunit surveillance complex called Cascade (CRISPR-associated complex for antiviral defence), which is required for protection against bacteriophages. Here we use cryo-electron microscopy to determine the subnanometre structures of Cascade before and after binding to a target sequence. These structures reveal a sea-horse-shaped architecture in which the crRNA is displayed along a helical arrangement of protein subunits that protect the crRNA from degradation while maintaining its availability for base pairing. Cascade engages invading nucleic acids through high-affinity base-pairing interactions near the 5' end of the crRNA. Base pairing extends along the crRNA, resulting in a series of short helical segments that trigger a concerted conformational change. This conformational rearrangement may serve as a signal that recruits a trans-acting nuclease (Cas3) for destruction of invading nucleic-acid sequences. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5314.map.gz emd_5314.map.gz | 10.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5314-v30.xml emd-5314-v30.xml emd-5314.xml emd-5314.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5314_1.png emd_5314_1.png | 172.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5314 http://ftp.pdbj.org/pub/emdb/structures/EMD-5314 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5314 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5314 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5314.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5314.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | reconstructed density of the E. coli cascade complex at 8 Angstroms resolution | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : E. coli Cascade complex
| Entire | Name: E. coli Cascade complex |
|---|---|
| Components |
|
-Supramolecule #1000: E. coli Cascade complex
| Supramolecule | Name: E. coli Cascade complex / type: sample / ID: 1000 / Details: The sample was monodisperse. / Oligomeric state: one asymmetric complex / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 405 KDa / Theoretical: 405 KDa / Method: Theoretical |
-Supramolecule #1: Cascade
| Supramolecule | Name: Cascade / type: organelle_or_cellular_component / ID: 1 / Name.synonym: Cascade / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Ref GO | 0: GO:0051607 |
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 405 KDa / Theoretical: 405 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 25 mM HEPES, 100 mM KCl, 1 mM TCEP |
| Grid | Details: 200 mesh Cu grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 78 K / Instrument: OTHER Details: Vitrification instrument: Vitrobot. blotting at 4 degrees C Method: 4 microliter aliquot of purified sample placed onto C-flat that had been glow-discharged in a nitrogen atmosphere for 60 sec using an Edwards Carbon Evaporator. The grids were blotted for 3 ...Method: 4 microliter aliquot of purified sample placed onto C-flat that had been glow-discharged in a nitrogen atmosphere for 60 sec using an Edwards Carbon Evaporator. The grids were blotted for 3 seconds using a blotting offset of -1. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 78 K / Max: 78 K / Average: 78 K |
| Date | Sep 17, 2010 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (4k x 4k) / Number real images: 2370 / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 100000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | From an initial set of 498137 automatically selected particles. Pre-processed with Appion. |
|---|---|
| CTF correction | Details: whole micrograph |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 8.8 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN2 SPARX / Number images used: 275573 |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















