[English] 日本語
 Yorodumi
Yorodumi- EMDB-5212: Structural insights into anti-parallel microtubule crosslinking b... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5212 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural insights into anti-parallel microtubule crosslinking by PRC1, a conserved non-motor microtubule associated protein. | |||||||||
 Map data Map data | This is a 3D map of a truncated version of PRCI | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | member of MAP65 family | |||||||||
| Biological species | unidentified (others) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 30.0 Å | |||||||||
 Authors Authors | Suramanian R / Wilson-Kubalek EM / Arthur CP / Bick MJ / Campbell EA / Darst SA / Milligan RA / Kapoor TM | |||||||||
 Citation Citation |  Journal: Cell / Year: 2010 Journal: Cell / Year: 2010Title: Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Authors: Radhika Subramanian / Elizabeth M Wilson-Kubalek / Christopher P Arthur / Matthew J Bick / Elizabeth A Campbell / Seth A Darst / Ronald A Milligan / Tarun M Kapoor /  Abstract: Formation of microtubule architectures, required for cell shape maintenance in yeast, directional cell expansion in plants and cytokinesis in eukaryotes, depends on antiparallel microtubule ...Formation of microtubule architectures, required for cell shape maintenance in yeast, directional cell expansion in plants and cytokinesis in eukaryotes, depends on antiparallel microtubule crosslinking by the conserved MAP65 protein family. Here, we combine structural and single molecule fluorescence methods to examine how PRC1, the human MAP65, crosslinks antiparallel microtubules. We find that PRC1's microtubule binding is mediated by a structured domain with a spectrin-fold and an unstructured Lys/Arg-rich domain. These two domains, at each end of a homodimer, are connected by a linkage that is flexible on single microtubules, but forms well-defined crossbridges between antiparallel filaments. Further, we show that PRC1 crosslinks are compliant and do not substantially resist filament sliding by motor proteins in vitro. Together, our data show how MAP65s, by combining structural flexibility and rigidity, tune microtubule associations to establish crosslinks that selectively "mark" antiparallel overlap in dynamic cytoskeletal networks. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5212.map.gz emd_5212.map.gz | 1.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5212-v30.xml emd-5212-v30.xml emd-5212.xml emd-5212.xml | 7.2 KB 7.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5212_1.png emd_5212_1.png | 258.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5212 http://ftp.pdbj.org/pub/emdb/structures/EMD-5212 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5212 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5212 | HTTPS FTP |
-Validation report
| Summary document |  emd_5212_validation.pdf.gz emd_5212_validation.pdf.gz | 76.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5212_full_validation.pdf.gz emd_5212_full_validation.pdf.gz | 75.9 KB | Display | |
| Data in XML |  emd_5212_validation.xml.gz emd_5212_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5212 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5212 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5212 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5212 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5212.map.gz / Format: CCP4 / Size: 11.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5212.map.gz / Format: CCP4 / Size: 11.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a 3D map of a truncated version of PRCI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
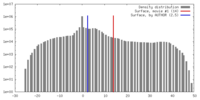
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Truncated form of PRC1
| Entire | Name: Truncated form of PRC1 |
|---|---|
| Components |
|
-Supramolecule #1000: Truncated form of PRC1
| Supramolecule | Name: Truncated form of PRC1 / type: sample / ID: 1000 / Details: complex of microtubule and protein / Number unique components: 2 |
|---|
-Macromolecule #1: microtubule
| Macromolecule | Name: microtubule / type: protein_or_peptide / ID: 1 / Name.synonym: tubulin / Recombinant expression: Yes / Database: NCBI |
|---|---|
| Source (natural) | Organism: unidentified (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: manual |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: gatan side entry / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
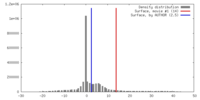
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 30.0 Å / Resolution method: OTHER / Software - Name: phoelix |
|---|---|
| CTF correction | Details: phoelix |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)