+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5198 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
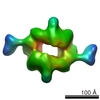
| Title | 3D reconstruction of negatively stained human raptor | |||||||||
 Map data Map data | Density map of human raptor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | metabolic role | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 28.0 Å | |||||||||
 Authors Authors | Yip CK / Murata K / Walz T / Sabatini DM / Kang SA | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2010 Journal: Mol Cell / Year: 2010Title: Structure of the human mTOR complex I and its implications for rapamycin inhibition. Authors: Calvin K Yip / Kazuyoshi Murata / Thomas Walz / David M Sabatini / Seong A Kang /  Abstract: The mammalian target of rapamycin complex 1 (mTORC1) regulates cell growth in response to the nutrient and energy status of the cell, and its deregulation is common in human cancers. Little is known ...The mammalian target of rapamycin complex 1 (mTORC1) regulates cell growth in response to the nutrient and energy status of the cell, and its deregulation is common in human cancers. Little is known about the overall architecture and subunit organization of this essential signaling complex. We have determined the three-dimensional (3D) structure of the fully assembled human mTORC1 by cryo-electron microscopy (cryo-EM). Our analyses reveal that mTORC1 is an obligate dimer with an overall rhomboid shape and a central cavity. The dimeric interfaces are formed by interlocking interactions between the mTOR and raptor subunits. Extended incubation with FKBP12-rapamycin compromises the structural integrity of mTORC1 in a stepwise manner, leading us to propose a model in which rapamycin inhibits mTORC1-mediated phosphorylation of 4E-BP1 and S6K1 through different mechanisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5198.map.gz emd_5198.map.gz | 392.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5198-v30.xml emd-5198-v30.xml emd-5198.xml emd-5198.xml | 9.3 KB 9.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5198_1.tif emd_5198_1.tif | 755.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5198 http://ftp.pdbj.org/pub/emdb/structures/EMD-5198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5198 | HTTPS FTP |
-Validation report
| Summary document |  emd_5198_validation.pdf.gz emd_5198_validation.pdf.gz | 78.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5198_full_validation.pdf.gz emd_5198_full_validation.pdf.gz | 77.9 KB | Display | |
| Data in XML |  emd_5198_validation.xml.gz emd_5198_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5198 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5198 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5198 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5198 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5198.map.gz / Format: CCP4 / Size: 825.2 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5198.map.gz / Format: CCP4 / Size: 825.2 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density map of human raptor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.48 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Purified human raptor
| Entire | Name: Purified human raptor |
|---|---|
| Components |
|
-Supramolecule #1000: Purified human raptor
| Supramolecule | Name: Purified human raptor / type: sample / ID: 1000 / Oligomeric state: monomer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: raptor
| Macromolecule | Name: raptor / type: protein_or_peptide / ID: 1 / Name.synonym: raptor / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human / Cell: HEK-293T / Location in cell: cytoplasm Homo sapiens (human) / synonym: human / Cell: HEK-293T / Location in cell: cytoplasm |
| Molecular weight | Theoretical: 150 KDa |
| Recombinant expression | Organism: Mammalian cells |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 50mM HEPES, 150mM NaCl |
| Staining | Type: NEGATIVE Details: protein solution was diluted 5x with buffer and adsorbed onto grids for 30 seconds before staining with 0.75% uranyl formate |
| Grid | Details: 400 mesh copper |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Date | Dec 1, 2007 |
| Image recording | Category: FILM / Film or detector model: GENERIC IMAGE PLATES / Digitization - Scanner: OTHER / Digitization - Sampling interval: 50 µm / Number real images: 83 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 67000 |
| Sample stage | Specimen holder: side entry room temperature holder / Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 60 |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















