+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of mouse PMCA captured in E2-P state (BEF3) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Calcium pump / Ptype-Atpase / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationP-type calcium transporter activity involved in regulation of postsynaptic cytosolic calcium ion concentration / cerebellar Purkinje cell layer morphogenesis / otolith mineralization / inner ear receptor cell differentiation / Reduction of cytosolic Ca++ levels / cGMP metabolic process / Ion transport by P-type ATPases / cerebellar granule cell differentiation / calcium-dependent ATPase activity / cerebellar Purkinje cell differentiation ...P-type calcium transporter activity involved in regulation of postsynaptic cytosolic calcium ion concentration / cerebellar Purkinje cell layer morphogenesis / otolith mineralization / inner ear receptor cell differentiation / Reduction of cytosolic Ca++ levels / cGMP metabolic process / Ion transport by P-type ATPases / cerebellar granule cell differentiation / calcium-dependent ATPase activity / cerebellar Purkinje cell differentiation / photoreceptor ribbon synapse / P-type Ca2+ transporter / P-type calcium transporter activity / detection of mechanical stimulus involved in sensory perception of sound / Ion homeostasis / positive regulation of calcium ion transport / serotonin metabolic process / auditory receptor cell stereocilium organization / locomotion / dendritic spine membrane / inner ear morphogenesis / neuromuscular process controlling balance / neuronal cell body membrane / regulation of cell size / parallel fiber to Purkinje cell synapse / inner ear development / cochlea development / glutamate receptor binding / regulation of cytosolic calcium ion concentration / lactation / presynaptic active zone membrane / cerebellum development / PDZ domain binding / locomotory behavior / sensory perception of sound / synapse organization / regulation of synaptic plasticity / postsynaptic density membrane / GABA-ergic synapse / cell morphogenesis / neuron differentiation / intracellular calcium ion homeostasis / calcium ion transport / presynaptic membrane / basolateral plasma membrane / postsynaptic membrane / calmodulin binding / apical plasma membrane / cilium / neuronal cell body / calcium ion binding / dendrite / glutamatergic synapse / endoplasmic reticulum / ATP hydrolysis activity / ATP binding / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.52 Å | |||||||||
 Authors Authors | Vinayagam D / Raunser S / Sistel O / Schulte U / Constantin CE / Prumbaum D / Zolles G / Fakler B | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2025 Journal: Nature / Year: 2025Title: Molecular mechanism of ultrafast transport by plasma membrane Ca-ATPases. Authors: Deivanayagabarathy Vinayagam / Oleg Sitsel / Uwe Schulte / Cristina E Constantin / Wout Oosterheert / Daniel Prumbaum / Gerd Zolles / Bernd Fakler / Stefan Raunser /   Abstract: Tight control of intracellular Ca levels is fundamental as they are used to control numerous signal transduction pathways. Plasma membrane Ca-ATPases (PMCAs) have a crucial role in this process by ...Tight control of intracellular Ca levels is fundamental as they are used to control numerous signal transduction pathways. Plasma membrane Ca-ATPases (PMCAs) have a crucial role in this process by extruding Ca against a steep concentration gradient from the cytosol to the extracellular space. Although new details of PMCA biology are constantly being uncovered, the structural basis of the most distinguishing features of these pumps, namely, transport rates in the kilohertz range and regulation of activity by the plasma membrane phospholipid PtdIns(4,5)P, has so far remained elusive. Here we present the structures of mouse PMCA2 in the presence and absence of its accessory subunit neuroplastin in eight different stages of its transport cycle. Combined with whole-cell recordings that accurately track PMCA-mediated Ca extrusion in intact cells, these structures enable us to establish the first comprehensive transport model for a PMCA, reveal the role of disease-causing mutations and uncover the structural underpinnings of regulatory PMCA-phospholipid interaction. The transport cycle-dependent dynamics of PtdIns(4,5)P are fundamental for its role as a 'latch' promoting the fast release of Ca and opening a passageway for counter-ions. These actions are required for maintaining the ultra-fast transport cycle. Moreover, we identify the PtdIns(4,5)P-binding site as an unanticipated target for drug-mediated manipulation of intracellular Ca levels. Our work provides detailed structural insights into the uniquely fast operation of native PMCA-type Ca pumps and its control by membrane lipids and drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_51558.map.gz emd_51558.map.gz | 97 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-51558-v30.xml emd-51558-v30.xml emd-51558.xml emd-51558.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_51558.png emd_51558.png | 89.6 KB | ||
| Filedesc metadata |  emd-51558.cif.gz emd-51558.cif.gz | 7.8 KB | ||
| Others |  emd_51558_half_map_1.map.gz emd_51558_half_map_1.map.gz emd_51558_half_map_2.map.gz emd_51558_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-51558 http://ftp.pdbj.org/pub/emdb/structures/EMD-51558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51558 | HTTPS FTP |
-Related structure data
| Related structure data |  9gsyMC  9gsdC  9gseC  9gsfC  9gsgC  9gshC  9gsiC  9gtbC  9gtiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_51558.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_51558.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_51558_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_51558_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mouse PMCA captured in E2-P state (BEF3)
| Entire | Name: mouse PMCA captured in E2-P state (BEF3) |
|---|---|
| Components |
|
-Supramolecule #1: mouse PMCA captured in E2-P state (BEF3)
| Supramolecule | Name: mouse PMCA captured in E2-P state (BEF3) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: prepared as separate cDNAs for transient (co)transfection of tsA201 nptn/basi double KO cells |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: Plasma membrane calcium-transporting ATPase 2
| Macromolecule | Name: Plasma membrane calcium-transporting ATPase 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: P-type Ca2+ transporter |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 134.647141 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGDMTNSDFY SKNQRNESSH GGEFGCTMEE LRSLMELRGT EAVVKIKETY GDTEAICRRL KTSPVEGLPG TAPDLEKRKQ IFGQNFIPP KKPKTFLQLV WEALQDVTLI ILEIAAIISL GLSFYHPPGE SNEGCATAQG GAEDEGEAEA GWIEGAAILL S VICVVLVT ...String: MGDMTNSDFY SKNQRNESSH GGEFGCTMEE LRSLMELRGT EAVVKIKETY GDTEAICRRL KTSPVEGLPG TAPDLEKRKQ IFGQNFIPP KKPKTFLQLV WEALQDVTLI ILEIAAIISL GLSFYHPPGE SNEGCATAQG GAEDEGEAEA GWIEGAAILL S VICVVLVT AFNDWSKEKQ FRGLQSRIEQ EQKFTVVRAG QVVQIPVAEI VVGDIAQIKY GDLLPADGLF IQGNDLKIDE SS LTGESDQ VRKSVDKDPM LLSGTHVMEG SGRMVVTAVG VNSQTGIIFT LLGAGGEEEE KKDKKAKQQD GAAAMEMQPL KSA EGGDAD DKKKANMHKK EKSVLQGKLT KLAVQIGKAG LVMSAITVII LVLYFTVDTF VVNKKPWLTE CTPVYVQYFV KFFI IGVTV LVVAVPEGLP LAVTISLAYS VKKMMKDNNL VRHLDACETM GNATAICSDK TGTLTTNRMT VVQAYVGDVH YKEIP DPSS INAKTLELLV NAIAINSAYT TKILPPEKEG ALPRQVGNKT ECGLLGFVLD LRQDYEPVRS QMPEEKLYKV YTFNSV RKS MSTVIKMPDE SFRMYSKGAS EIVLKKCCKI LSGAGEARVF RPRDRDEMVK KVIEPMACDG LRTICVAYRD FPSSPEP DW DNENDILNEL TCICVVGIED PVRPEVPEAI RKCQRAGITV RMVTGDNINT ARAIAIKCGI IHPGEDFLCL EGKEFNRR I RNEKGEIEQE RIDKIWPKLR VLARSSPTDK HTLVKGIIDS THTEQRQVVA VTGDGTNDGP ALKKADVGFA MGIAGTDVA KEASDIILTD DNFSSIVKAV MWGRNVYDSI SKFLQFQLTV NVVAVIVAFT GACITQDSPL KAVQMLWVNL IMDTFASLAL ATEPPTETL LLRKPYGRNK PLISRTMMKN ILGHAVYQLT LIFTLLFVGE KMFQIDSGRN APLHSPPSEH YTIIFNTFVM M QLFNEINA RKIHGERNVF DGIFRNPIFC TIVLGTFAIQ IVIVQFGGKP FSCSPLQLDQ WMWCIFIGLG ELVWGQVIAT IP TSRLKFL KEAGRLTQKE EIPEEELNED VEEIDHAERE LRRGQILWFR GLNRIQTQIR VVKAFRSSLY EGLEKPESRT SIH NFMAHP EFRIEDSQPH IPLIDDTDLE EDAALKQNSS PPSSLNKNNS AIDSGINLTT DTSKSATSSS PGSPIHSLET SLEN LYFQG GDYKDDDDK UniProtKB: Plasma membrane calcium-transporting ATPase 2 |
-Macromolecule #2: BERYLLIUM TRIFLUORIDE ION
| Macromolecule | Name: BERYLLIUM TRIFLUORIDE ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: BEF |
|---|---|
| Molecular weight | Theoretical: 66.007 Da |
| Chemical component information |  ChemComp-BEF: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bi...
| Macromolecule | Name: (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate type: ligand / ID: 4 / Number of copies: 1 / Formula: KXP |
|---|---|
| Molecular weight | Theoretical: 1.047088 KDa |
| Chemical component information | 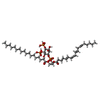 ChemComp-KXP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: Tris 20mM NaCl 150mM |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | The complex was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Temperature | Min: 93.0 K / Max: 113.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 13134 / Average exposure time: 3.5 sec. / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 55000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































