+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Befiradol-bound serotonin 5-HT1A receptor - Gs Protein Complex | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Complex / Serotonin / Befiradol / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of serotonin secretion / Gi/o-coupled serotonin receptor activity / regulation of hormone secretion / regulation of behavior / receptor-receptor interaction / Serotonin receptors / regulation of dopamine metabolic process / serotonin receptor activity / G protein-coupled serotonin receptor activity / serotonin receptor signaling pathway ...regulation of serotonin secretion / Gi/o-coupled serotonin receptor activity / regulation of hormone secretion / regulation of behavior / receptor-receptor interaction / Serotonin receptors / regulation of dopamine metabolic process / serotonin receptor activity / G protein-coupled serotonin receptor activity / serotonin receptor signaling pathway / neurotransmitter receptor activity / serotonin metabolic process / serotonin binding / gamma-aminobutyric acid signaling pathway / exploration behavior / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / D1 dopamine receptor binding / intracellular transport / regulation of vasoconstriction / vascular endothelial cell response to laminar fluid shear stress / behavioral fear response / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / adenylate cyclase-inhibiting serotonin receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / bone development / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / chemical synaptic transmission / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / positive regulation of cell population proliferation / synapse / dendrite Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Schneider J / Gmeiner P / Boettcher B / Rasmussen T | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2025 Journal: Sci Adv / Year: 2025Title: Discovery of a functionally selective serotonin receptor (5-HTR) agonist for the treatment of pain. Authors: Annika Ullrich / Johannes Schneider / João M Braz / Eduard Neu / Nico Staffen / Markus Stanek / Jana Bláhová / Tamsanqa Hove / Tamara Albert / Anni Allikalt / Stefan Löber / Karnika ...Authors: Annika Ullrich / Johannes Schneider / João M Braz / Eduard Neu / Nico Staffen / Markus Stanek / Jana Bláhová / Tamsanqa Hove / Tamara Albert / Anni Allikalt / Stefan Löber / Karnika Bhardwaj / Sian Rodriguez-Rosado / Elissa Fink / Tim Rasmussen / Harald Hübner / Asuka Inoue / Brian K Shoichet / Allan I Basbaum / Bettina Böttcher / Dorothee Weikert / Peter Gmeiner /    Abstract: The heterotrimeric G protein-coupled serotonin receptor 5-HT receptor (5-HTR) mediates antinociception and may serve as a valuable target for the treatment of pain. Starting from a chemical library, ...The heterotrimeric G protein-coupled serotonin receptor 5-HT receptor (5-HTR) mediates antinociception and may serve as a valuable target for the treatment of pain. Starting from a chemical library, we evolved ST171, a bitopic 5-HTR agonist that revealed highly potent and functionally selective G signaling without G activation and marginal β-arrestin recruitment. ST171 is effective in acute and chronic pain models. Cryo-electron microscopy structures of ST171 bound to 5-HTR in complex with the G protein compared to the canonical agonist befiradol bound to complexes of 5-HTR with G or G revealed that the ligands occupy different exo-sites. The individual binding poses are associated with ligand-specific receptor conformations that were further studied by molecular dynamics simulations, allowing us to better understand ligand bias, a phenomenon that may be crucial to the discovery of more effective and safe G protein-coupled receptor drugs. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_51434.map.gz emd_51434.map.gz | 77.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-51434-v30.xml emd-51434-v30.xml emd-51434.xml emd-51434.xml | 25.9 KB 25.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_51434_fsc.xml emd_51434_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_51434.png emd_51434.png | 46.6 KB | ||
| Filedesc metadata |  emd-51434.cif.gz emd-51434.cif.gz | 7.8 KB | ||
| Others |  emd_51434_half_map_1.map.gz emd_51434_half_map_1.map.gz emd_51434_half_map_2.map.gz emd_51434_half_map_2.map.gz | 65.5 MB 65.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-51434 http://ftp.pdbj.org/pub/emdb/structures/EMD-51434 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51434 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51434 | HTTPS FTP |
-Related structure data
| Related structure data |  9gl2MC  8pjkC  8pkmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_51434.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_51434.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.946 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_51434_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_51434_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tern
| Entire | Name: Tern |
|---|---|
| Components |
|
-Supramolecule #1: Tern
| Supramolecule | Name: Tern / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.683434 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQA DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ AALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENIRRVF NDCRDIIQRM HLRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.810352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHG SSGSELDQLR QEAEQLKNQI RDARKACADA TLSQITNNID PVGRIQMRTR RTLRGHLAKI YAMHWGTDSR LLVSASQDG KLIIWDSYTT NKVHAIPLRS SWVMTCAYAP SGNYVACGGL DNICSIYNLK TREGNVRVSR ELAGHTGYLS C CRFLDDNQ ...String: MHHHHHHHHG SSGSELDQLR QEAEQLKNQI RDARKACADA TLSQITNNID PVGRIQMRTR RTLRGHLAKI YAMHWGTDSR LLVSASQDG KLIIWDSYTT NKVHAIPLRS SWVMTCAYAP SGNYVACGGL DNICSIYNLK TREGNVRVSR ELAGHTGYLS C CRFLDDNQ IVTSSGDTTC ALWDIETGQQ TTTFTGHTGD VMSLSLAPDT RLFVSGACDA SAKLWDVREG MCRQTFTGHE SD INAICFF PNGNAFATGS DDATCRLFDL RADQELMTYS HDNIICGITS VSFSKSGRLL LAGYDDFNCN VWDALKADRA GVL AGHDNR VSCLGVTDDG MAVATGSWDS FLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: 5-hydroxytryptamine receptor 1A
| Macromolecule | Name: 5-hydroxytryptamine receptor 1A / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.431578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAAAAHHH HHHHHHHENL YFQGMDVLSP GQGNNTTSPP APFETGGNTT GISDVTVSYQ VITSLLLGT LIFCAVLGNA CVVAAIALER SLQNVANYLI GSLAVTDLMV SVLVLPMAAL YQVLNKWTLG QVTCDLFIAL D VLCCTSSI ...String: MKTIIALSYI FCLVFADYKD DDDAAAAHHH HHHHHHHENL YFQGMDVLSP GQGNNTTSPP APFETGGNTT GISDVTVSYQ VITSLLLGT LIFCAVLGNA CVVAAIALER SLQNVANYLI GSLAVTDLMV SVLVLPMAAL YQVLNKWTLG QVTCDLFIAL D VLCCTSSI WHLCAIALDR YWAITDPIDY VNKRTPRRAA ALISLTWLIG FLISIPPMLG WRTPEDRSDP DACTISKDHG YT IYSTFGA FYIPLLLMLV LYGRIFRAAR FRIRKTVKKV EKTGADTRHG ASPAPQPKKS VNGESGSRNW RLGVESKAGG ALC ANGAVR QGDDGAALEV IEVHRVGNSK EHLPLPSEAG PTPCAPASFE RKNERNAEAK RKMALARERK TVKTLGIIMG TFIL CWLPF FIVALVLPFC ESSCHMPTLL GAIINWLGYS NSLLNPVIYA YFNKDFQNAF KKIIKCKFCR Q UniProtKB: 5-hydroxytryptamine receptor 1A |
-Macromolecule #5: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 5 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #6: (2R)-1-(heptadecanoyloxy)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,...
| Macromolecule | Name: (2R)-1-(heptadecanoyloxy)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate type: ligand / ID: 6 / Number of copies: 1 / Formula: T7M |
|---|---|
| Molecular weight | Theoretical: 953.081 Da |
| Chemical component information | 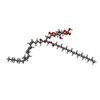 ChemComp-T7M: |
-Macromolecule #7: (3-chloranyl-4-fluoranyl-phenyl)-[4-fluoranyl-4-[[(5-methylpyridi...
| Macromolecule | Name: (3-chloranyl-4-fluoranyl-phenyl)-[4-fluoranyl-4-[[(5-methylpyridin-2-yl)methylamino]methyl]piperidin-1-yl]methanone type: ligand / ID: 7 / Number of copies: 1 / Formula: ZKV |
|---|---|
| Molecular weight | Theoretical: 393.858 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 120 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 5 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 8000 / Average exposure time: 5.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)





































