+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of CyclinB1 N-terminus bound to the NCP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Arginine anchor / NCP / Cyclin B1 / Complex / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to DDT / cyclin B1-CDK1 complex / positive regulation of mitochondrial ATP synthesis coupled electron transport / regulation of chromosome condensation / Mitotic Prophase / E2F-enabled inhibition of pre-replication complex formation / ventricular cardiac muscle cell development / Depolymerization of the Nuclear Lamina / MASTL Facilitates Mitotic Progression / positive regulation of mRNA 3'-end processing ...response to DDT / cyclin B1-CDK1 complex / positive regulation of mitochondrial ATP synthesis coupled electron transport / regulation of chromosome condensation / Mitotic Prophase / E2F-enabled inhibition of pre-replication complex formation / ventricular cardiac muscle cell development / Depolymerization of the Nuclear Lamina / MASTL Facilitates Mitotic Progression / positive regulation of mRNA 3'-end processing / regulation of mitotic cell cycle spindle assembly checkpoint / positive regulation of attachment of spindle microtubules to kinetochore / Activation of NIMA Kinases NEK9, NEK6, NEK7 / patched binding / Phosphorylation of Emi1 / Transcriptional regulation by RUNX2 / Nuclear Pore Complex (NPC) Disassembly / Phosphorylation of the APC/C / G2/M DNA replication checkpoint / tissue regeneration / outer kinetochore / Initiation of Nuclear Envelope (NE) Reformation / Polo-like kinase mediated events / Golgi Cisternae Pericentriolar Stack Reorganization / cellular response to fatty acid / cyclin-dependent protein serine/threonine kinase activator activity / nucleosome disassembly / oocyte maturation / digestive tract development / Condensation of Prometaphase Chromosomes / UV-damage excision repair / cyclin-dependent protein serine/threonine kinase regulator activity / mitotic metaphase chromosome alignment / Regulation of APC/C activators between G1/S and early anaphase / ubiquitin-like protein ligase binding / microtubule organizing center / positive regulation of G2/M transition of mitotic cell cycle / response to mechanical stimulus / Regulation of MITF-M-dependent genes involved in cell cycle and proliferation / positive regulation of cardiac muscle cell proliferation / Cyclin A/B1/B2 associated events during G2/M transition / negative regulation of megakaryocyte differentiation / Chk1/Chk2(Cds1) mediated inactivation of Cyclin B:Cdk1 complex / protein localization to CENP-A containing chromatin / Chromatin modifying enzymes / Replacement of protamines by nucleosomes in the male pronucleus / Nuclear events stimulated by ALK signaling in cancer / CENP-A containing nucleosome / Packaging Of Telomere Ends / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / Deposition of new CENPA-containing nucleosomes at the centromere / positive regulation of mitotic cell cycle / APC/C:Cdc20 mediated degradation of Cyclin B / telomere organization / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / Interleukin-7 signaling / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest / epigenetic regulation of gene expression / RNA Polymerase I Promoter Opening / Inhibition of DNA recombination at telomere / Assembly of the ORC complex at the origin of replication / Meiotic synapsis / Resolution of Sister Chromatid Cohesion / SUMOylation of chromatin organization proteins / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / DNA methylation / Condensation of Prophase Chromosomes / Chromatin modifications during the maternal to zygotic transition (MZT) / mitotic spindle organization / SIRT1 negatively regulates rRNA expression / HCMV Late Events / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / Defective pyroptosis / Negative Regulation of CDH1 Gene Transcription / HDACs deacetylate histones / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / cellular response to iron(III) ion / Nonhomologous End-Joining (NHEJ) / RNA Polymerase I Promoter Escape / Transcriptional regulation by small RNAs / G1/S transition of mitotic cell cycle / Formation of the beta-catenin:TCF transactivating complex / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / HDMs demethylate histones / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / G2/M DNA damage checkpoint / NoRC negatively regulates rRNA expression / B-WICH complex positively regulates rRNA expression / PKMTs methylate histone lysines / DNA Damage/Telomere Stress Induced Senescence / Pre-NOTCH Transcription and Translation / Meiotic recombination / G2/M transition of mitotic cell cycle / Activation of anterior HOX genes in hindbrain development during early embryogenesis / Metalloprotease DUBs / positive regulation of fibroblast proliferation Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) / | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Young RVC / Muhammad R / Alfieri C | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation | Journal: Acta Crystallogr D Struct Biol / Year: 2018 Title: Real-space refinement in PHENIX for cryo-EM and crystallography. Authors: Pavel V Afonine / Billy K Poon / Randy J Read / Oleg V Sobolev / Thomas C Terwilliger / Alexandre Urzhumtsev / Paul D Adams /    Abstract: This article describes the implementation of real-space refinement in the phenix.real_space_refine program from the PHENIX suite. The use of a simplified refinement target function enables very fast ...This article describes the implementation of real-space refinement in the phenix.real_space_refine program from the PHENIX suite. The use of a simplified refinement target function enables very fast calculation, which in turn makes it possible to identify optimal data-restraint weights as part of routine refinements with little runtime cost. Refinement of atomic models against low-resolution data benefits from the inclusion of as much additional information as is available. In addition to standard restraints on covalent geometry, phenix.real_space_refine makes use of extra information such as secondary-structure and rotamer-specific restraints, as well as restraints or constraints on internal molecular symmetry. The re-refinement of 385 cryo-EM-derived models available in the Protein Data Bank at resolutions of 6 Å or better shows significant improvement of the models and of the fit of these models to the target maps. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50443.map.gz emd_50443.map.gz | 6.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50443-v30.xml emd-50443-v30.xml emd-50443.xml emd-50443.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_50443.png emd_50443.png | 109.6 KB | ||
| Filedesc metadata |  emd-50443.cif.gz emd-50443.cif.gz | 7 KB | ||
| Others |  emd_50443_half_map_1.map.gz emd_50443_half_map_1.map.gz emd_50443_half_map_2.map.gz emd_50443_half_map_2.map.gz | 80.9 MB 80.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50443 http://ftp.pdbj.org/pub/emdb/structures/EMD-50443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50443 | HTTPS FTP |
-Related structure data
| Related structure data |  9fh9MC  9fgqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50443.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50443.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.94 Å | ||||||||||||||||||||||||||||||||||||
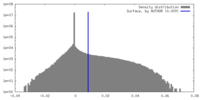
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_50443_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
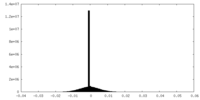
| Density Histograms |
-Half map: #1
| File | emd_50443_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
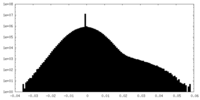
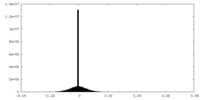
| Density Histograms |
- Sample components
Sample components
-Entire : Cyclin B1 NTD bound to the acidic path of the NCP
| Entire | Name: Cyclin B1 NTD bound to the acidic path of the NCP |
|---|---|
| Components |
|
-Supramolecule #1: Cyclin B1 NTD bound to the acidic path of the NCP
| Supramolecule | Name: Cyclin B1 NTD bound to the acidic path of the NCP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: G2/mitotic-specific cyclin-B1
| Macromolecule | Name: G2/mitotic-specific cyclin-B1 / type: protein_or_peptide / ID: 1 / Details: The first 21 amino acids of cyclin B1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.407878 KDa |
| Sequence | String: MALRVTRNSK INAENKAKIN M UniProtKB: G2/mitotic-specific cyclin-B1 |
-Macromolecule #2: Histone H3.1
| Macromolecule | Name: Histone H3.1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.437167 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PATGGVKKPH RYRPGTVALR EIRRYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSSA VMALQEACEA YLVGLFEDTN LCAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3.1 |
-Macromolecule #3: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.394426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-Macromolecule #4: Histone H2A type 3
| Macromolecule | Name: Histone H2A type 3 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.151523 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKQGGK ARAKAKSRSS RAGLQFPVGR VHRLLRKGNY SERVGAGAPV YLAAVLEYLT AEILELAGNA ARDNKKTRII PRHLQLAIR NDEELNKLLG RVTIAQGGVL PNIQAVLLPK KTESHHKAKG K UniProtKB: Histone H2A type 3 |
-Macromolecule #5: Histone H2B 1.1
| Macromolecule | Name: Histone H2B 1.1 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.655948 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKSAPAPKK GSKKAVTKTQ KKDGKKRRKT RKESYAIYVY KVLKQVHPDT GISSKAMSIM NSFVNDVFER IAGEASRLAH YNKRSTITS REIQTAVRLL LPGELAKHAV SEGTKAVTKY TSAK UniProtKB: Histone H2B 1.1 |
-Macromolecule #6: DNA (145-MER)
| Macromolecule | Name: DNA (145-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.13877 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT) ...String: (DA)(DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT) (DA)(DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT) (DA)(DA) (DA)(DC)(DG)(DC)(DA)(DC)(DG) (DT)(DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT) (DC)(DC)(DC) (DC)(DC)(DG)(DC)(DG)(DT) (DT)(DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC) (DA)(DA)(DG)(DG) (DG)(DG)(DA)(DT)(DT) (DA)(DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT) (DC)(DT)(DC)(DC)(DA) (DG)(DG)(DC)(DA) (DC)(DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DC) (DA)(DT)(DC) (DC)(DG)(DA)(DT) |
-Macromolecule #7: DNA (145-MER)
| Macromolecule | Name: DNA (145-MER) / type: dna / ID: 7 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.610043 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC) ...String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT) (DA)(DA) (DA)(DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC) (DG)(DT)(DA) (DC)(DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC) (DA)(DA)(DT)(DT)(DG) (DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC) (DC)(DG)(DG)(DG)(DA)(DT) (DT)(DC)(DT) (DC)(DG)(DA)(DT) |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 5 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 66184 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)