+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human APC3loop 375-381 bound to the NCP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Arginine anchor / NCP / APC3 / Complex / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationConversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase / Inactivation of APC/C via direct inhibition of the APC/C complex / APC/C:Cdc20 mediated degradation of mitotic proteins / anaphase-promoting complex / regulation of meiotic cell cycle / Aberrant regulation of mitotic exit in cancer due to RB1 defects / anaphase-promoting complex-dependent catabolic process / protein branched polyubiquitination / metaphase/anaphase transition of mitotic cell cycle / Phosphorylation of the APC/C ...Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase / Inactivation of APC/C via direct inhibition of the APC/C complex / APC/C:Cdc20 mediated degradation of mitotic proteins / anaphase-promoting complex / regulation of meiotic cell cycle / Aberrant regulation of mitotic exit in cancer due to RB1 defects / anaphase-promoting complex-dependent catabolic process / protein branched polyubiquitination / metaphase/anaphase transition of mitotic cell cycle / Phosphorylation of the APC/C / protein K11-linked ubiquitination / Regulation of APC/C activators between G1/S and early anaphase / Transcriptional Regulation by VENTX / protein K48-linked ubiquitination / negative regulation of megakaryocyte differentiation / protein localization to CENP-A containing chromatin / Chromatin modifying enzymes / Replacement of protamines by nucleosomes in the male pronucleus / CENP-A containing nucleosome / Packaging Of Telomere Ends / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / Deposition of new CENPA-containing nucleosomes at the centromere / APC/C:Cdc20 mediated degradation of Cyclin B / telomere organization / regulation of mitotic cell cycle / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / APC-Cdc20 mediated degradation of Nek2A / Interleukin-7 signaling / epigenetic regulation of gene expression / RNA Polymerase I Promoter Opening / Inhibition of DNA recombination at telomere / Assembly of the ORC complex at the origin of replication / Meiotic synapsis / SUMOylation of chromatin organization proteins / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / DNA methylation / Condensation of Prophase Chromosomes / Chromatin modifications during the maternal to zygotic transition (MZT) / SIRT1 negatively regulates rRNA expression / HCMV Late Events / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Autodegradation of Cdh1 by Cdh1:APC/C / Regulation of endogenous retroelements by KRAB-ZFP proteins / APC/C:Cdc20 mediated degradation of Securin / Defective pyroptosis / Negative Regulation of CDH1 Gene Transcription / HDACs deacetylate histones / Assembly of the pre-replicative complex / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Nonhomologous End-Joining (NHEJ) / RNA Polymerase I Promoter Escape / Transcriptional regulation by small RNAs / Formation of the beta-catenin:TCF transactivating complex / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / HDMs demethylate histones / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 / G2/M DNA damage checkpoint / NoRC negatively regulates rRNA expression / B-WICH complex positively regulates rRNA expression / PKMTs methylate histone lysines / DNA Damage/Telomere Stress Induced Senescence / CDK-mediated phosphorylation and removal of Cdc6 / Pre-NOTCH Transcription and Translation / Meiotic recombination / spindle / Activation of anterior HOX genes in hindbrain development during early embryogenesis / Metalloprotease DUBs / Transcriptional regulation of granulopoiesis / RMTs methylate histone arginines / neuron projection development / HCMV Early Events / mitotic spindle / structural constituent of chromatin / Separation of Sister Chromatids / UCH proteinases / heterochromatin formation / nucleosome / Antigen processing: Ubiquitination & Proteasome degradation / nucleosome assembly / E3 ubiquitin ligases ubiquitinate target proteins / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / HATs acetylate histones / Factors involved in megakaryocyte development and platelet production / RUNX1 regulates transcription of genes involved in differentiation of HSCs / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / chromatin organization / Processing of DNA double-strand break ends / Senescence-Associated Secretory Phenotype (SASP) / protein phosphatase binding / Oxidative Stress Induced Senescence / Estrogen-dependent gene expression / chromosome, telomeric region / Ub-specific processing proteases / protein ubiquitination / cadherin binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Young RVC / Muhammad R / Alfieri C | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2024 Journal: EMBO J / Year: 2024Title: Spatial control of the APC/C ensures the rapid degradation of cyclin B1. Authors: Luca Cirillo / Rose Young / Sapthaswaran Veerapathiran / Annalisa Roberti / Molly Martin / Azzah Abubacar / Camilla Perosa / Catherine Coates / Reyhan Muhammad / Theodoros I Roumeliotis / ...Authors: Luca Cirillo / Rose Young / Sapthaswaran Veerapathiran / Annalisa Roberti / Molly Martin / Azzah Abubacar / Camilla Perosa / Catherine Coates / Reyhan Muhammad / Theodoros I Roumeliotis / Jyoti S Choudhary / Claudio Alfieri / Jonathon Pines /  Abstract: The proper control of mitosis depends on the ubiquitin-mediated degradation of the right mitotic regulator at the right time. This is effected by the Anaphase Promoting Complex/Cyclosome (APC/C) ...The proper control of mitosis depends on the ubiquitin-mediated degradation of the right mitotic regulator at the right time. This is effected by the Anaphase Promoting Complex/Cyclosome (APC/C) ubiquitin ligase that is regulated by the Spindle Assembly Checkpoint (SAC). The SAC prevents the APC/C from recognising Cyclin B1, the essential anaphase and cytokinesis inhibitor, until all chromosomes are attached to the spindle. Once chromosomes are attached, Cyclin B1 is rapidly degraded to enable chromosome segregation and cytokinesis. We have a good understanding of how the SAC inhibits the APC/C, but relatively little is known about how the APC/C recognises Cyclin B1 as soon as the SAC is turned off. Here, by combining live-cell imaging, in vitro reconstitution biochemistry, and structural analysis by cryo-electron microscopy, we provide evidence that the rapid recognition of Cyclin B1 in metaphase requires spatial regulation of the APC/C. Using fluorescence cross-correlation spectroscopy, we find that Cyclin B1 and the APC/C primarily interact at the mitotic apparatus. We show that this is because Cyclin B1, like the APC/C, binds to nucleosomes, and identify an 'arginine-anchor' in the N-terminus as necessary and sufficient for binding to the nucleosome. Mutating the arginine anchor on Cyclin B1 reduces its interaction with the APC/C and delays its degradation: cells with the mutant, non-nucleosome-binding Cyclin B1 become aneuploid, demonstrating the physiological relevance of our findings. Together, our data demonstrate that mitotic chromosomes promote the efficient interaction between Cyclin B1 and the APC/C to ensure the timely degradation of Cyclin B1 and genomic stability. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50416.map.gz emd_50416.map.gz | 4.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50416-v30.xml emd-50416-v30.xml emd-50416.xml emd-50416.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_50416.png emd_50416.png | 126.8 KB | ||
| Filedesc metadata |  emd-50416.cif.gz emd-50416.cif.gz | 7.6 KB | ||
| Others |  emd_50416_half_map_1.map.gz emd_50416_half_map_1.map.gz emd_50416_half_map_2.map.gz emd_50416_half_map_2.map.gz | 29.7 MB 29.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50416 http://ftp.pdbj.org/pub/emdb/structures/EMD-50416 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50416 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50416 | HTTPS FTP |
-Related structure data
| Related structure data |  9fgqMC  9fh9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50416.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50416.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
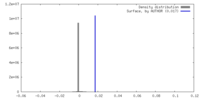
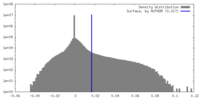
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_50416_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
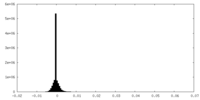
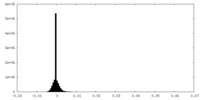
| Density Histograms |
-Half map: #2
| File | emd_50416_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
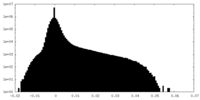
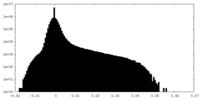
| Density Histograms |
- Sample components
Sample components
-Entire : APC3 motif bound to the NCP acidic patch
| Entire | Name: APC3 motif bound to the NCP acidic patch |
|---|---|
| Components |
|
-Supramolecule #1: APC3 motif bound to the NCP acidic patch
| Supramolecule | Name: APC3 motif bound to the NCP acidic patch / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3, #6-#7 Details: Recombinant protein sample of residues 375-381 of APC3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Cell division cycle protein 27 homolog
| Macromolecule | Name: Cell division cycle protein 27 homolog / type: protein_or_peptide / ID: 1 Details: This is a disorder loop of human APC3 residues 177-446 fused to a SpyTag via a 27 residue GSA linker. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.788488 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GGSASNCLPN SCTTQVPNHS LSHRQPETVL TETPQDTIEL NRLNLESSNS KYSLNTDSSV SYIDSAVISP DTVPLGTGTS ILSKQVQNK PKTGRSLLGG PAALSPLTPS FGILPLETPS PGDGSYLQNY TNTPPVIDVP STGAPSKKSV ARIGQTGTKS V FSQSGNSR ...String: GGSASNCLPN SCTTQVPNHS LSHRQPETVL TETPQDTIEL NRLNLESSNS KYSLNTDSSV SYIDSAVISP DTVPLGTGTS ILSKQVQNK PKTGRSLLGG PAALSPLTPS FGILPLETPS PGDGSYLQNY TNTPPVIDVP STGAPSKKSV ARIGQTGTKS V FSQSGNSR EVTPILAQTQ SSGPQTSTTP QVLSPTITSP PNALPRRSSR LFTSDSSTTK ENSKKLKMKF PPKIPNRKTK SK TNKGGIT QPNINDSLEI TKLDSSIISE GKISTITGSA GSAGSAGSAG SAGSAGSAGS AGSARGVPHI VMVDAYKRYK UniProtKB: Cell division cycle protein 27 homolog |
-Macromolecule #2: Histone H3.1
| Macromolecule | Name: Histone H3.1 / type: protein_or_peptide / ID: 2 / Details: Human histone H3.1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.437167 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PATGGVKKPH RYRPGTVALR EIRRYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSSA VMALQEACEA YLVGLFEDTN LCAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3.1 |
-Macromolecule #3: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 3 / Details: Human histone H4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.394426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-Macromolecule #4: Histone H2A type 2-A
| Macromolecule | Name: Histone H2A type 2-A / type: protein_or_peptide / ID: 4 Details: This is a H2A/H2B fusion protein with a SpyCatcher tag attached Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.125549 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKQGGK ARAKAKSRSS RAGLQFPVGR VHRLLRKGNY AERVGAGAPV YMAAVLEYLT AEILELAGNA ARDNKKTRII PRHLQLAIR NDEELNKLLG KVTIAQGGVL PNIQAVLLPK KTESHHKAKG K UniProtKB: Histone H2A type 2-A |
-Macromolecule #5: Histone H2B type 1-B
| Macromolecule | Name: Histone H2B type 1-B / type: protein_or_peptide / ID: 5 Details: This is a H2A/H2B fusion protein with a SpyCatcher tag attached,This is a H2A/H2B fusion protein with a SpyCatcher tag attached Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.445771 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VTTLSGLSGE QGPSGDMTTE EDSATHIKFS KRDEDGRELA GATMELRDSS GKTISTWISD GHVKDFYLYP GKYTFVETAA PDGYEVATP IEFTVNEDGQ VTVDGEATEG DAHTGSAWSH PQFEKGSAGS AAGSGAGWSH PQFEKGSAMP EPSKSAPAPK K GSKKAITK ...String: VTTLSGLSGE QGPSGDMTTE EDSATHIKFS KRDEDGRELA GATMELRDSS GKTISTWISD GHVKDFYLYP GKYTFVETAA PDGYEVATP IEFTVNEDGQ VTVDGEATEG DAHTGSAWSH PQFEKGSAGS AAGSGAGWSH PQFEKGSAMP EPSKSAPAPK K GSKKAITK AQKKDGKKRK RSRKESYSIY VYKVLKQVHP DTGISSKAMG IMNSFVNDIF ERIAGEASRL AHYNKRSTIT SR EIQTAVR LLLPGELAKH AVSEGTKAVT KYTSSK UniProtKB: Histone H2B type 1-B |
-Macromolecule #6: DNA (132-MER)
| Macromolecule | Name: DNA (132-MER) / type: dna / ID: 6 Details: The Widom 147 bp sequence with 32 nucleotides of DNA on either side Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 64.911336 KDa |
| Sequence | String: (DA)(DT)(DC)(DT)(DT)(DA)(DG)(DC)(DG)(DC) (DG)(DG)(DT)(DG)(DA)(DG)(DT)(DT)(DC)(DA) (DA)(DA)(DT)(DA)(DC)(DC)(DC)(DG)(DG) (DC)(DA)(DA)(DA)(DT)(DC)(DG)(DA)(DG)(DA) (DA) (DT)(DC)(DC)(DC)(DG)(DG) ...String: (DA)(DT)(DC)(DT)(DT)(DA)(DG)(DC)(DG)(DC) (DG)(DG)(DT)(DG)(DA)(DG)(DT)(DT)(DC)(DA) (DA)(DA)(DT)(DA)(DC)(DC)(DC)(DG)(DG) (DC)(DA)(DA)(DA)(DT)(DC)(DG)(DA)(DG)(DA) (DA) (DT)(DC)(DC)(DC)(DG)(DG)(DT)(DG) (DC)(DC)(DG)(DA)(DG)(DG)(DC)(DC)(DG)(DC) (DT)(DC) (DA)(DA)(DT)(DT)(DG)(DG)(DT) (DC)(DG)(DT)(DA)(DG)(DA)(DC)(DA)(DG)(DC) (DT)(DC)(DT) (DA)(DG)(DC)(DA)(DC)(DC) (DG)(DC)(DT)(DT)(DA)(DA)(DA)(DC)(DG)(DC) (DA)(DC)(DG)(DT) (DA)(DC)(DG)(DC)(DG) (DC)(DT)(DG)(DT)(DC)(DC)(DC)(DC)(DC)(DG) (DC)(DG)(DT)(DT)(DT) (DT)(DA)(DA)(DC) (DC)(DG)(DC)(DC)(DA)(DA)(DG)(DG)(DG)(DG) (DA)(DT)(DT)(DA)(DC)(DT) (DC)(DC)(DC) (DT)(DA)(DG)(DT)(DC)(DT)(DC)(DC)(DA)(DG) (DG)(DC)(DA)(DC)(DG)(DT)(DG) (DT)(DC) (DA)(DG)(DA)(DT)(DA)(DT)(DA)(DT)(DA)(DC) (DA)(DT)(DC)(DC)(DG)(DA)(DT)(DT) (DT) (DG)(DC)(DC)(DG)(DG)(DG)(DT)(DA)(DT)(DT) (DT)(DG)(DA)(DA)(DC)(DT)(DC)(DA)(DC) (DC)(DG)(DC)(DG)(DC)(DT)(DA)(DA)(DG)(DA) (DT) |
-Macromolecule #7: DNA (131-MER)
| Macromolecule | Name: DNA (131-MER) / type: dna / ID: 7 Details: Widom 147 DNA sequence flanked with 32 nucleotides on either side Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 65.382633 KDa |
| Sequence | String: (DA)(DT)(DC)(DT)(DT)(DA)(DG)(DC)(DG)(DC) (DG)(DG)(DT)(DG)(DA)(DG)(DT)(DT)(DC)(DA) (DA)(DA)(DT)(DA)(DC)(DC)(DC)(DG)(DG) (DC)(DA)(DA)(DA)(DT)(DC)(DG)(DG)(DA)(DT) (DG) (DT)(DA)(DT)(DA)(DT)(DA) ...String: (DA)(DT)(DC)(DT)(DT)(DA)(DG)(DC)(DG)(DC) (DG)(DG)(DT)(DG)(DA)(DG)(DT)(DT)(DC)(DA) (DA)(DA)(DT)(DA)(DC)(DC)(DC)(DG)(DG) (DC)(DA)(DA)(DA)(DT)(DC)(DG)(DG)(DA)(DT) (DG) (DT)(DA)(DT)(DA)(DT)(DA)(DT)(DC) (DT)(DG)(DA)(DC)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT) (DG)(DG)(DA)(DG)(DA)(DC)(DT) (DA)(DG)(DG)(DG)(DA)(DG)(DT)(DA)(DA)(DT) (DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DC) (DG)(DG)(DT)(DT)(DA)(DA)(DA)(DA)(DC)(DG) (DC)(DG)(DG)(DG) (DG)(DG)(DA)(DC)(DA) (DG)(DC)(DG)(DC)(DG)(DT)(DA)(DC)(DG)(DT) (DG)(DC)(DG)(DT)(DT) (DT)(DA)(DA)(DG) (DC)(DG)(DG)(DT)(DG)(DC)(DT)(DA)(DG)(DA) (DG)(DC)(DT)(DG)(DT)(DC) (DT)(DA)(DC) (DG)(DA)(DC)(DC)(DA)(DA)(DT)(DT)(DG)(DA) (DG)(DC)(DG)(DG)(DC)(DC)(DT) (DC)(DG) (DG)(DC)(DA)(DC)(DC)(DG)(DG)(DG)(DA)(DT) (DT)(DC)(DT)(DC)(DG)(DA)(DT)(DT) (DT) (DG)(DC)(DC)(DG)(DG)(DG)(DT)(DA)(DT)(DT) (DT)(DG)(DA)(DA)(DC)(DT)(DC)(DA)(DC) (DC)(DG)(DC)(DG)(DC)(DT)(DA)(DA)(DG)(DA) (DT) |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 75 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
Details: 20 mM HEPEs pH8.0, 50 mM NaCl, 0.5 mM TCEP | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)