[English] 日本語
 Yorodumi
Yorodumi- EMDB-50424: SARS-CoV-2 (B.1.1.529/Omicron variant) Spike protein in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
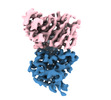
| Title | SARS-CoV-2 (B.1.1.529/Omicron variant) Spike protein in complex with the single chain fragment scFv76 (focused refinement) | |||||||||
 Map data Map data | sharped map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / Spike / single chain fragment / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvirion component / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...virion component / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Berlinguer M / Chaves-Sanjuan A / Milazzo FM / Minenkova O / De Santis R / Bolognesi M | |||||||||
| Funding support |  Italy, 1 items Italy, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of scFv76 in complex with SARS-CoV-2 Omicron Spike protein Authors: Chaves-Sanjuan A / Bolognesi M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50424.map.gz emd_50424.map.gz | 229.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50424-v30.xml emd-50424-v30.xml emd-50424.xml emd-50424.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_50424.png emd_50424.png | 82 KB | ||
| Masks |  emd_50424_msk_1.map emd_50424_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50424.cif.gz emd-50424.cif.gz | 7.1 KB | ||
| Others |  emd_50424_additional_1.map.gz emd_50424_additional_1.map.gz emd_50424_half_map_1.map.gz emd_50424_half_map_1.map.gz emd_50424_half_map_2.map.gz emd_50424_half_map_2.map.gz | 120.6 MB 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50424 http://ftp.pdbj.org/pub/emdb/structures/EMD-50424 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50424 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50424 | HTTPS FTP |
-Related structure data
| Related structure data |  9fgtMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50424.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50424.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharped map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.889 Å | ||||||||||||||||||||||||||||||||||||
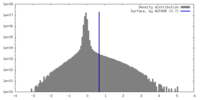
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50424_msk_1.map emd_50424_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
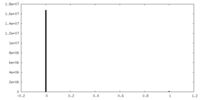
| Density Histograms |
-Additional map: unsharped map
| File | emd_50424_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharped map | ||||||||||||
| Projections & Slices |
| ||||||||||||
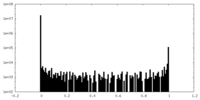
| Density Histograms |
-Half map: #2
| File | emd_50424_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
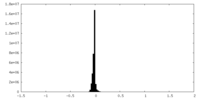
| Density Histograms |
-Half map: #1
| File | emd_50424_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Spike protein in complex with the single chain fragmen...
| Entire | Name: SARS-CoV-2 Spike protein in complex with the single chain fragment scFv76 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Spike protein in complex with the single chain fragmen...
| Supramolecule | Name: SARS-CoV-2 Spike protein in complex with the single chain fragment scFv76 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Molecular weight | Theoretical: 510 KDa |
-Supramolecule #2: SARS-CoV-2 Spike protein
| Supramolecule | Name: SARS-CoV-2 Spike protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: scFv76 single chain fragment
| Supramolecule | Name: scFv76 single chain fragment / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein,Fibritin
| Macromolecule | Name: Spike glycoprotein,Fibritin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) |
| Molecular weight | Theoretical: 135.255156 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISR FDNPVLPFND GVYFASIEK SNIIRGWIFG TTLDSKTQSL LIVNNATNVV IKVCEFQFCN DPFLDVYYHK NNKSWMESEF RVYSSANNCT F EYVSQPFL ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISR FDNPVLPFND GVYFASIEK SNIIRGWIFG TTLDSKTQSL LIVNNATNVV IKVCEFQFCN DPFLDVYYHK NNKSWMESEF RVYSSANNCT F EYVSQPFL MDLEGKQGNF KNLREFVFKN IDGYFKIYSK HTPIIVPEDL PQGFSALEPL VDLPIGINIT RFQTLLALHR SS SGWTAGA AAYYVGYLQP RTFLLKYNEN GTITDAVDCA LDPLSETKCT LKSFTVEKGI YQTSNFRVQP TESIVRFPNI TNL CPFDEV FNATRFASVY AWNRKRISNC VADYSVLYNL APFFTFKCYG VSPTKLNDLC FTNVYADSFV IRGDEVRQIA PGQT GNIAD YNYKLPDDFT GCVIAWNSNK LDSKVSGNYN YLYRLFRKSN LKPFERDIST EIYQAGNKPC NGVAGFNCYF PLRSY SFRP TYGVGHQPYR VVVLSFELLH APATVCGPKK STNLVKNKCV NFNFNGLKGT GVLTESNKKF LPFQQFGRDI ADTTDA VRD PQTLEILDIT PCSFGGVSVI TPGTNTSNQV AVLYQGVNCT EVNVFQTRAG CLIGAEYVNN SYECDIPIGA GICASYQ TS QSIIAYTMSL GAENSVAYSN NSIAIPTNFT ISVTTEILPV SMTKTSVDCT MYICGDSTEC SNLLLQYGSF CTQLKRAL T GIAVEQDKNT QEVFAQVKQI YKTPPIKYFG GFNFSQILPD PSKSKRSFIE DLLFNKVTKF KGLTVLPPLL TDEMIAQYT SALLAGTITS GWTFGAGAAL QIPFAMQMAY RFNGIGVTQN VLYENQKLIA NQFNSAIGKI QDSLSSTASA LGKLQDVVNH NAQALNTLV KQLSSKFGAI SSVLNDIFSR LDPPEAEVQI DRLITGRLQS LQTYVTQQLI RAAEIRASAN LAATKMSECV L GQSKRVDF CGKGYHLMSF PQSAPHGVVF LHVTYVPAQE KNFTTAPAIC HDGKAHFPRE GVFVSNGTHW FVTQRNFYEP QI ITTDNTF VSGNCDVVIG IVNNTVYDPL QPELDSFKEE LDKYFKNHTS PDVDLGDISG INASVVNIQK EIDRLNEVAK NLN ESLIDL QELGKYEQGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGRS LEVLFQGPGH HHHHHHHSAW SHPQFEKGGG SGGG GSGGS AWSHPQFEK UniProtKB: Spike glycoprotein, Fibritin |
-Macromolecule #2: Single chain fragment scFv76
| Macromolecule | Name: Single chain fragment scFv76 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.040947 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: RTMEEVQLLQ SAGGLVQPGG SLRLSCAASG FTVSANYMSW VRQAPGKGLE WVSVIYPGGS TFYADSVKGR FTISRDNSKN TLYLQMNSL RVEDTAVYYC ARDLSVAGAF DIWGQGTLVT VSSGGGGSGG GGSGGGGSEI VLTQSPGTLS LSPGERATLS C RASQSVSS ...String: RTMEEVQLLQ SAGGLVQPGG SLRLSCAASG FTVSANYMSW VRQAPGKGLE WVSVIYPGGS TFYADSVKGR FTISRDNSKN TLYLQMNSL RVEDTAVYYC ARDLSVAGAF DIWGQGTLVT VSSGGGGSGG GGSGGGGSEI VLTQSPGTLS LSPGERATLS C RASQSVSS SYLAWYQQKP GQAPRLLIYG ASSRATGIPD RFSGSGSGTD FTLTISRLEP EDFAVYYCQQ YGSSPYTFGQ GT KLEIKRA AAGDYKDDDD KHHHHHH |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: PBS |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 7179 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)