[English] 日本語
 Yorodumi
Yorodumi- EMDB-5038: 11-Angstrom cryo-EM reconstruction of nucleotide-free Nod complex... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5038 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 11-Angstrom cryo-EM reconstruction of nucleotide-free Nod complexed to the 13-protofilament microtubule. | |||||||||
 Map data Map data | 11-Angstrom cryo-EM reconstruction of nucleotide-free Nod complexed to the 13-protofilament microtubule. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | kinesin / tubulin / nod / motor proteins / cytoskeletal motor | |||||||||
| Function / homology |  Function and homology information Function and homology informationestablishment of meiotic spindle orientation / Hedgehog 'on' state / distributive segregation / COPI-dependent Golgi-to-ER retrograde traffic / Kinesins / meiotic chromosome segregation / female meiotic nuclear division / spindle assembly involved in female meiosis I / microtubule plus-end binding / positive regulation of axon guidance ...establishment of meiotic spindle orientation / Hedgehog 'on' state / distributive segregation / COPI-dependent Golgi-to-ER retrograde traffic / Kinesins / meiotic chromosome segregation / female meiotic nuclear division / spindle assembly involved in female meiosis I / microtubule plus-end binding / positive regulation of axon guidance / kinesin complex / microtubule motor activity / tubulin complex / microtubule-based movement / microtubule-based process / cytoplasmic microtubule / positive regulation of microtubule polymerization / cellular response to interleukin-4 / structural constituent of cytoskeleton / microtubule cytoskeleton organization / neuron migration / mitotic cell cycle / double-stranded RNA binding / microtubule cytoskeleton / microtubule binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / cilium / protein heterodimerization activity / GTPase activity / ubiquitin protein ligase binding / GTP binding / ATP hydrolysis activity / ATP binding / metal ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species | unidentified (others) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 11.0 Å | |||||||||
 Authors Authors | Cochran JC / Sindelar CV / Mulko NK / Collins KA / Kong SE / Hawley RS / Kull FJ | |||||||||
 Citation Citation |  Journal: Cell / Year: 2009 Journal: Cell / Year: 2009Title: ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement. Authors: Jared C Cochran / Charles V Sindelar / Natasha K Mulko / Kimberly A Collins / Stephanie E Kong / R Scott Hawley / F Jon Kull /  Abstract: Segregation of nonexchange chromosomes during Drosophila melanogaster meiosis requires the proper function of NOD, a nonmotile kinesin-10. We have determined the X-ray crystal structure of the NOD ...Segregation of nonexchange chromosomes during Drosophila melanogaster meiosis requires the proper function of NOD, a nonmotile kinesin-10. We have determined the X-ray crystal structure of the NOD catalytic domain in the ADP- and AMPPNP-bound states. These structures reveal an alternate conformation of the microtubule binding region as well as a nucleotide-sensitive relay of hydrogen bonds at the active site. Additionally, a cryo-electron microscopy reconstruction of the nucleotide-free microtubule-NOD complex shows an atypical binding orientation. Thermodynamic studies show that NOD binds tightly to microtubules in the nucleotide-free state, yet other nucleotide states, including AMPPNP, are weakened. Our pre-steady-state kinetic analysis demonstrates that NOD interaction with microtubules occurs slowly with weak activation of ADP product release. Upon rapid substrate binding, NOD detaches from the microtubule prior to the rate-limiting step of ATP hydrolysis, which is also atypical for a kinesin. We propose a model for NOD's microtubule plus-end tracking that drives chromosome movement. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5038.map.gz emd_5038.map.gz | 20.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5038-v30.xml emd-5038-v30.xml emd-5038.xml emd-5038.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5038.jpg emd_5038.jpg | 155.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5038 http://ftp.pdbj.org/pub/emdb/structures/EMD-5038 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5038 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5038 | HTTPS FTP |
-Related structure data
| Related structure data |  3dcoMC  3dc4C  3dcbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5038.map.gz / Format: CCP4 / Size: 21.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5038.map.gz / Format: CCP4 / Size: 21.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 11-Angstrom cryo-EM reconstruction of nucleotide-free Nod complexed to the 13-protofilament microtubule. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Nucleotide-free Nod complexed to the 13-protofilament microtubule
| Entire | Name: Nucleotide-free Nod complexed to the 13-protofilament microtubule |
|---|---|
| Components |
|
-Supramolecule #1000: Nucleotide-free Nod complexed to the 13-protofilament microtubule
| Supramolecule | Name: Nucleotide-free Nod complexed to the 13-protofilament microtubule type: sample / ID: 1000 Oligomeric state: One monomer of Nod binds to one heterodimer of tubulin Number unique components: 3 |
|---|---|
| Molecular weight | Theoretical: 164 KDa Method: Theoretical MW of one Nod monomer complexed to one tubulin dimer, based on length of the protein chains. |
-Macromolecule #1: Nod
| Macromolecule | Name: Nod / type: protein_or_peptide / ID: 1 / Name.synonym: Nod / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism: unidentified (others) / Strain: B834(DE3) / Location in cell: cytosol |
| Molecular weight | Theoretical: 43 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #2: microtubule
| Macromolecule | Name: microtubule / type: protein_or_peptide / ID: 2 / Name.synonym: tubulin / Oligomeric state: quasi-helical assembly / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism: unidentified (others) / Tissue: brain / Cell: cow / Location in cell: cytoplasm |
| Molecular weight | Theoretical: 120 KDa |
| Sequence | GO: tubulin complex / InterPro: Alpha tubulin, Beta tubulin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 / Details: see publication |
|---|---|
| Grid | Details: 300 mesh copper grid with homemade holey carbon film |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 103 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: homemade. Carried out under ambient conditions. No glow discharge was applied to the grids prior to sample application. Method: Excess solution wicked away from grid with filter paper before blotting and plunging immediately. Less than 0.5 seconds elapsed between blotting and plunging. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 4000EX |
|---|---|
| Temperature | Min: 98 K / Max: 108 K / Average: 100 K |
| Alignment procedure | Legacy - Astigmatism: crude, software correction used |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.3 µm / Number real images: 130 / Average electron dose: 16 e/Å2 Details: Data from only 17 micrographs were selected for the final reconstruction. Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 400 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.1 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 11.0 Å / Resolution method: OTHER / Software - Name: SPIDER Details: Image defocus and astigmatism were determined by ctffind3. |
|---|---|
| CTF correction | Details: Integrated with fourier inversion (similar to FREALIGN) |
| Final angle assignment | Details: SPIDER |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: SITUS |
| Details | Protocol: Rigid Body. Because Nod occupancy was low, most tubulin density was deleted using UCSF Chimera"s volume eraser prior to automatic fitting to avoid spurious fits. |
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation |
| Output model |  PDB-3dco: |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name: SITUS |
| Details | Protocol: Rigid Body |
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation |
| Output model |  PDB-3dco: |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera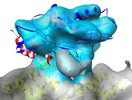













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















