[English] 日本語
 Yorodumi
Yorodumi- EMDB-50252: Human choline transporter-like protein 1 (hCTL1/SLC44A1) in LMNG -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human choline transporter-like protein 1 (hCTL1/SLC44A1) in LMNG | |||||||||
 Map data Map data | local refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Choline transporter-like family / CTL / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationethanolamine transport / ethanolamine transmembrane transporter activity / Transport of bile salts and organic acids, metal ions and amine compounds / Choline catabolism / choline catabolic process / choline transmembrane transporter activity / phosphatidylcholine biosynthetic process / choline transport / antiporter activity / Synthesis of PC ...ethanolamine transport / ethanolamine transmembrane transporter activity / Transport of bile salts and organic acids, metal ions and amine compounds / Choline catabolism / choline catabolic process / choline transmembrane transporter activity / phosphatidylcholine biosynthetic process / choline transport / antiporter activity / Synthesis of PC / transport across blood-brain barrier / transmembrane transport / mitochondrial outer membrane / mitochondrion / extracellular exosome / nucleoplasm / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
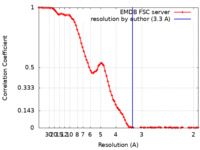
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Driller JH / Pedersen BP | |||||||||
| Funding support |  Denmark, European Union, 2 items Denmark, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: To be announced Authors: Driller JH / Pedersen BP | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50252.map.gz emd_50252.map.gz | 26.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50252-v30.xml emd-50252-v30.xml emd-50252.xml emd-50252.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50252_fsc.xml emd_50252_fsc.xml | 11 KB | Display |  FSC data file FSC data file |
| Images |  emd_50252.png emd_50252.png | 78.3 KB | ||
| Masks |  emd_50252_msk_1.map emd_50252_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50252.cif.gz emd-50252.cif.gz | 5.3 KB | ||
| Others |  emd_50252_additional_1.map.gz emd_50252_additional_1.map.gz emd_50252_additional_2.map.gz emd_50252_additional_2.map.gz emd_50252_half_map_1.map.gz emd_50252_half_map_1.map.gz emd_50252_half_map_2.map.gz emd_50252_half_map_2.map.gz | 49.7 MB 26.3 MB 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50252 http://ftp.pdbj.org/pub/emdb/structures/EMD-50252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50252 | HTTPS FTP |
-Validation report
| Summary document |  emd_50252_validation.pdf.gz emd_50252_validation.pdf.gz | 956 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50252_full_validation.pdf.gz emd_50252_full_validation.pdf.gz | 955.6 KB | Display | |
| Data in XML |  emd_50252_validation.xml.gz emd_50252_validation.xml.gz | 15.4 KB | Display | |
| Data in CIF |  emd_50252_validation.cif.gz emd_50252_validation.cif.gz | 19.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50252 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50252 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50252 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50252 | HTTPS FTP |
-Related structure data
| Related structure data |  9qu3M  5o7zC  5o80C  5o81C  5o82C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50252.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50252.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9705 Å | ||||||||||||||||||||||||||||||||||||
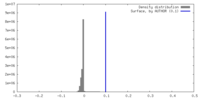
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50252_msk_1.map emd_50252_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
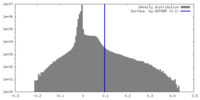
| Density Histograms |
-Additional map: sharpened map
| File | emd_50252_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
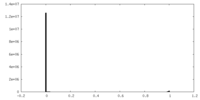
| Density Histograms |
-Additional map: local refinement map with improved density for the...
| File | emd_50252_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local refinement map with improved density for the extracellular domain | ||||||||||||
| Projections & Slices |
| ||||||||||||
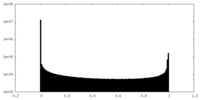
| Density Histograms |
-Half map: #1
| File | emd_50252_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_50252_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Choline transporter-like protein 1 in LMNG
| Entire | Name: Choline transporter-like protein 1 in LMNG |
|---|---|
| Components |
|
-Supramolecule #1: Choline transporter-like protein 1 in LMNG
| Supramolecule | Name: Choline transporter-like protein 1 in LMNG / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all / Details: choline transporter-like protein 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: human choline transporter-like protein 1
| Macromolecule | Name: human choline transporter-like protein 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MGCCSSASSA AQSSKREWKP LEDRSCTDIP WLLLFILFCI GMGFICGFSI ATGAAARLVS GYDSYGNICG QKNTKLEAIP NSGMDHTQRK YVFFLDPCNL DLINRKIKSV ALCVAACPRQ ELKTLSDVQK FAEINGSALC SYNLKPSEYT TSPKSSVLCP KLPVPASAPI ...String: MGCCSSASSA AQSSKREWKP LEDRSCTDIP WLLLFILFCI GMGFICGFSI ATGAAARLVS GYDSYGNICG QKNTKLEAIP NSGMDHTQRK YVFFLDPCNL DLINRKIKSV ALCVAACPRQ ELKTLSDVQK FAEINGSALC SYNLKPSEYT TSPKSSVLCP KLPVPASAPI PFFHRCAPVN ISCYAKFAEA LITFVSDNSV LHRLISGVMT SKEIILGLCL LSLVLSMILM VIIRYISRVL VWILTILVIL GSLGGTGVLW WLYAKQRRSP KETVTPEQLQ IAEDNLRALL IYAISATVFT VILFLIMLVM RKRVALTIAL FHVAGKVFIH LPLLVFQPFW TFFALVLFWV YWIMTLLFLG TTGSPVQNEQ GFVEFKISGP LQYMWWYHVV GLIWISEFIL ACQQMTVAGA VVTYYFTRDK RNLPFTPILA SVNRLIRYHL GTVAKGSFII TLVKIPRMIL MYIHSQLKGK ENACARCVLK SCICCLWCLE KCLNYLNQNA YTATAINSTN FCTSAKDAFV ILVENALRVA TINTVGDFML FLGKVLIVCS TGLAGIMLLN YQQDYTVWVL PLIIVCLFAF LVAHCFLSIY EMVVDVLFLC FAIDTKYNDG SPGREFYMDK VLMEFVENSR KAMKEAGKGG VADSRELKPM ASGASSALSA LVPR UniProtKB: Choline transporter-like protein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 6758 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)