[English] 日本語
 Yorodumi
Yorodumi- EMDB-5009: A cryo-EM map of the FimD-tip complex, a bacterial surface pilus ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5009 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
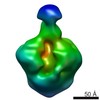
| Title | A cryo-EM map of the FimD-tip complex, a bacterial surface pilus assembly intermediate in complex with the outer membrane secretion channel. | |||||||||
 Map data Map data | 3D Cryo-EM map of FimD-tip complex, a bacterial outer membrane pilus assembly intermediate | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryo-electron microscopy / bacterial pilus / bacterial outer membrane secretion channel / pilus biogenesis | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 23.0 Å | |||||||||
 Authors Authors | Tang C / Thanassi D / Li H | |||||||||
 Citation Citation |  Journal: Cell / Year: 2008 Journal: Cell / Year: 2008Title: Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Authors: Han Remaut / Chunyan Tang / Nadine S Henderson / Jerome S Pinkner / Tao Wang / Scott J Hultgren / David G Thanassi / Gabriel Waksman / Huilin Li /  Abstract: Gram-negative pathogens commonly exhibit adhesive pili on their surfaces that mediate specific attachment to the host. A major class of pili is assembled via the chaperone/usher pathway. Here, the ...Gram-negative pathogens commonly exhibit adhesive pili on their surfaces that mediate specific attachment to the host. A major class of pili is assembled via the chaperone/usher pathway. Here, the structural basis for pilus fiber assembly and secretion performed by the outer membrane assembly platform--the usher--is revealed by the crystal structure of the translocation domain of the P pilus usher PapC and single particle cryo-electron microscopy imaging of the FimD usher bound to a translocating type 1 pilus assembly intermediate. These structures provide molecular snapshots of a twinned-pore translocation machinery in action. Unexpectedly, only one pore is used for secretion, while both usher protomers are used for chaperone-subunit complex recruitment. The translocating pore itself comprises 24 beta strands and is occluded by a folded plug domain, likely gated by a conformationally constrained beta-hairpin. These structures capture the secretion of a virulence factor across the outer membrane of gram-negative bacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5009.map.gz emd_5009.map.gz | 3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5009-v30.xml emd-5009-v30.xml emd-5009.xml emd-5009.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5009_1.tif emd_5009_1.tif | 750.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5009 http://ftp.pdbj.org/pub/emdb/structures/EMD-5009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5009 | HTTPS FTP |
-Validation report
| Summary document |  emd_5009_validation.pdf.gz emd_5009_validation.pdf.gz | 78.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5009_full_validation.pdf.gz emd_5009_full_validation.pdf.gz | 77.2 KB | Display | |
| Data in XML |  emd_5009_validation.xml.gz emd_5009_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5009 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5009 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5009 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5009 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5009.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5009.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D Cryo-EM map of FimD-tip complex, a bacterial outer membrane pilus assembly intermediate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.54 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : FimD-tip complex
| Entire | Name: FimD-tip complex |
|---|---|
| Components |
|
-Supramolecule #1000: FimD-tip complex
| Supramolecule | Name: FimD-tip complex / type: sample / ID: 1000 / Details: The sample was monodisperse Oligomeric state: FimD usher dimer in complex with one copy each of FimH, FimF, FimG pilins and FimC chaperone Number unique components: 5 |
|---|---|
| Molecular weight | Experimental: 260 KDa / Theoretical: 260 KDa |
-Supramolecule #1: FimD-tip complex
| Supramolecule | Name: FimD-tip complex / type: organelle_or_cellular_component / ID: 1 / Name.synonym: Pilus assembly usher / Details: This component forms a dimer in the complex / Number of copies: 1 / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Ref GO | 0: GO:0015473 |
| Ref INTERPRO | 0: IPR000015 |
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 96 KDa / Theoretical: 96 KDa |
| Recombinant expression | Organism: Escherichia coli B strain Tuner (Novagen) / Recombinant plasmid: Tuner/pAN2 and pNH237 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.04 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM Tris-HCl (pH 8), 0.15 M NaCl, 0.05% DDM. |
| Grid | Details: glow-discharged lacey carbon grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 108 K / Instrument: OTHER Details: Vitrification instrument: Vitrobot. 12 degree Celsius chamber temperature Method: 6 seconds blot |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Temperature | Min: 100 K / Max: 105 K / Average: 103 K |
| Alignment procedure | Legacy - Astigmatism: correction at 250,000 mag / Legacy - Electron beam tilt params: -2 mrad |
| Date | Feb 1, 2007 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 12.7 µm / Number real images: 100 / Average electron dose: 10 e/Å2 / Od range: 1.3 / Bits/pixel: 14 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Gatan 626 cryo holder / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | particles were manually selected |
|---|---|
| CTF correction | Details: Each films |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN, SPIDER / Number images used: 11000 |
| Final two d classification | Number classes: 100 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | PDBEntryID_givenInChain. Protocol: Rigid Body. manual docking followed by local correlation based real space fitting in chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: correlation |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















