+ Open data
Open data
 Loading...
Loading...
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the flowering plant mitoribosome with P-site tRNA | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Mitochondria / tRNA / mitoribosome / ribosome / plant | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationendosperm cellularization / pollen tube development / endosperm development / regulation of starch biosynthetic process / pollen germination / nucleus localization / pollen development / regulation of programmed cell death / embryo development ending in seed dormancy / cell communication ...endosperm cellularization / pollen tube development / endosperm development / regulation of starch biosynthetic process / pollen germination / nucleus localization / pollen development / regulation of programmed cell death / embryo development ending in seed dormancy / cell communication / vacuole organization / mitochondrial large ribosomal subunit / mitochondrial small ribosomal subunit / mitochondrial membrane organization / regulation of gene expression / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / mitochondrion / RNA binding Similarity search - Function Mitochondrial ribosomal protein S7, plants / Ribosomal protein S3, mitochondrial, plant / : / Eukaryotic mitochondrial regulator protein / Ribosomal protein S7, bacterial/organellar-type / Ribosomal protein S3, C-terminal / Ribosomal protein S3, C-terminal domain / Ribosomal protein S3, C-terminal domain superfamily / K homology domain superfamily, prokaryotic type / Ribosomal Proteins L2, RNA binding N-terminal domain ...Mitochondrial ribosomal protein S7, plants / Ribosomal protein S3, mitochondrial, plant / : / Eukaryotic mitochondrial regulator protein / Ribosomal protein S7, bacterial/organellar-type / Ribosomal protein S3, C-terminal / Ribosomal protein S3, C-terminal domain / Ribosomal protein S3, C-terminal domain superfamily / K homology domain superfamily, prokaryotic type / Ribosomal Proteins L2, RNA binding N-terminal domain / Ribosomal protein S5/S7 / Ribosomal protein S7 domain / Ribosomal protein S7 domain superfamily / Ribosomal protein S7p/S5e / Ribosomal Proteins L2, RNA binding domain / Ribosomal Proteins L2, RNA binding domain / Ribosomal protein L2 / Nucleic acid-binding, OB-fold Similarity search - Domain/homology Small ribosomal subunit protein uS3m / Small ribosomal subunit protein uS7m / Uncharacterized protein / Large ribosomal subunit protein uL2mz, N-terminal part Similarity search - Component | |||||||||||||||
| Biological species |    | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | |||||||||||||||
 Authors Authors | Waltz F / Skaltsogiannis V / Giege P | |||||||||||||||
| Funding support |  Germany, Germany,  Switzerland, 2 items Switzerland, 2 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structural insights into cauliflower mitoribosome in translation state and in association with a late assembly factor. Authors: Vasileios Skaltsogiannis / Philippe Wolff / Tan-Trung Nguyen / Nicolas Corre / David Pflieger / Todd Blevins / Yaser Hashem / Philippe Giegé / Florent Waltz /   Abstract: Ribosomes are key molecular machines that translate mRNA into proteins. Mitoribosomes are specific ribosomes found in mitochondria, which have been shown to be remarkably diverse across eukaryotic ...Ribosomes are key molecular machines that translate mRNA into proteins. Mitoribosomes are specific ribosomes found in mitochondria, which have been shown to be remarkably diverse across eukaryotic lineages. In plants, they possess unique features, including additional rRNA domains stabilized by plant-specific proteins. However, the structural specificities of plant mitoribosomes in translation state remained unknown. We used cryo-electron microscopy to provide a high-resolution structural characterization of the cauliflower mitoribosome, in translating and maturation states. The structure reveals the mitoribosome bound with a tRNA in the peptidyl site, along with a segment of mRNA and a nascent polypeptide. Moreover, using structural data, nanopore sequencing and mass spectrometry, we identify a set of 19 ribosomal RNA modifications. Additionally, we observe a late assembly intermediate of the small ribosomal subunit, in complex with the RsgA assembly factor. This reveals how a plant-specific extension of RsgA blocks the mRNA channel to prevent premature mRNA association before complete small subunit maturation. Our findings elucidate key aspects of translation in angiosperm plant mitochondria, revealing its distinct features compared to other eukaryotic lineages. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50011.map.gz emd_50011.map.gz | 646.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50011-v30.xml emd-50011-v30.xml emd-50011.xml emd-50011.xml | 115.2 KB 115.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50011_fsc.xml emd_50011_fsc.xml | 18.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_50011.png emd_50011.png | 111 KB | ||
| Masks |  emd_50011_msk_1.map emd_50011_msk_1.map | 684.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50011.cif.gz emd-50011.cif.gz | 24.8 KB | ||
| Others |  emd_50011_half_map_1.map.gz emd_50011_half_map_1.map.gz emd_50011_half_map_2.map.gz emd_50011_half_map_2.map.gz | 635.1 MB 635.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50011 http://ftp.pdbj.org/pub/emdb/structures/EMD-50011 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50011 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50011 | HTTPS FTP |
-Validation report
| Summary document |  emd_50011_validation.pdf.gz emd_50011_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50011_full_validation.pdf.gz emd_50011_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_50011_validation.xml.gz emd_50011_validation.xml.gz | 27.9 KB | Display | |
| Data in CIF |  emd_50011_validation.cif.gz emd_50011_validation.cif.gz | 37.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50011 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50011 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50011 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50011 | HTTPS FTP |
-Related structure data
| Related structure data |  9evsMC  9gytC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50011.map.gz / Format: CCP4 / Size: 684.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50011.map.gz / Format: CCP4 / Size: 684.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50011_msk_1.map emd_50011_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_50011_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_50011_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Flowering plant mitochondrial ribosome in presence of P-site tRNA
| Entire | Name: Flowering plant mitochondrial ribosome in presence of P-site tRNA |
|---|---|
| Components |
|
+Supramolecule #1: Flowering plant mitochondrial ribosome in presence of P-site tRNA
| Supramolecule | Name: Flowering plant mitochondrial ribosome in presence of P-site tRNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#88 |
|---|---|
| Source (natural) | Organism:  |
+Macromolecule #1: Small ribosomal subunit protein uS3m
| Macromolecule | Name: Small ribosomal subunit protein uS3m / type: protein_or_peptide / ID: 1 / Details: GWHPBJSH000741 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.731312 KDa |
| Sequence | String: MARKGNPISV RLGKNRSSDS SWFSDYYYGK FVYQDVNLRS YFGSIRPPTR LTFGFRLGRC ILLHFPKRTF IHFFLPRRPR RLKRREKTR PGKEKGRWWT TFGKAGPIGC LRDDTEEERN EVRGRGARKR VESIRLDDRK KQNEIRGWPK KKQRYGYHDR S PSIKKNLS ...String: MARKGNPISV RLGKNRSSDS SWFSDYYYGK FVYQDVNLRS YFGSIRPPTR LTFGFRLGRC ILLHFPKRTF IHFFLPRRPR RLKRREKTR PGKEKGRWWT TFGKAGPIGC LRDDTEEERN EVRGRGARKR VESIRLDDRK KQNEIRGWPK KKQRYGYHDR S PSIKKNLS KLLRISGAFK HPKYAGVVND IAFLIENDDS FKKTKLFKFF FPKKSRSDGP TSYLRTLPAV RPSLNFLVMQ YF FNTKNQM NFDPVVVLNH FVAPGAAEPS TMGRANAQGR SLQKRIRSRI AFFVESLTSE KKCLAEAKNR LTHFIRLAND LRF AGTTKT TISLFPFFGA TFFFLRDGVG VYNNLDAREQ LLNQLRVKCW NLLGKDKVME LIEKFKNLGG IEELIKVIDM MIEI ILRKR GIPYRYNSYF YEVKKMRSFL SNRTNTKTLI ESVKIKSVYQ SASLIAQDIS FQLKNKRRSF HSIFAKIVKE IPKRV EGIR ICFSGRLKDA AEKAQTKCYK HRKTSCNVFN QKIDYAPVEV STRYGILGVK VWISYSQKKG GRAISETYEI UniProtKB: Small ribosomal subunit protein uS3m |
+Macromolecule #2: uS4m
| Macromolecule | Name: uS4m / type: protein_or_peptide / ID: 2 / Details: GWHPBJSH000696 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.438488 KDa |
| Sequence | String: MWLLKKLIQR DIDMSPLRFQ TCRLLLGNVW NRELTIIQRR ILRRLRNRKR SIKKRKIYSK KYLTSYIQLQ TTRKLSLFYG DLPITEMHR GTKRTSYIPF LLNLETRFDV ILLRLHFLET IPQARQLISH RRVCVNKGMV SITHLKLSHG DIISFQENNA I IRGEEIRR ...String: MWLLKKLIQR DIDMSPLRFQ TCRLLLGNVW NRELTIIQRR ILRRLRNRKR SIKKRKIYSK KYLTSYIQLQ TTRKLSLFYG DLPITEMHR GTKRTSYIPF LLNLETRFDV ILLRLHFLET IPQARQLISH RRVCVNKGMV SITHLKLSHG DIISFQENNA I IRGEEIRR SFYKEILVEK IIGKLLHQPL RMWRRSKTEW FHLLKTKRGC RLLLKSRFLQ QLRSSMQEED LERTKKFGSE KV CLGSSFA EHKRMKRNLL KSLFLSKRRK EKNLNLPTRT ISPIVYNSSL SLYSNSTYCF ASPHKLTMKR RIKRIELPTH YLE VNYRTP KAVVFYGPNI GHIPHDIRLK DLNLLLWSRN GRGQNI |
+Macromolecule #3: uS5m
| Macromolecule | Name: uS5m / type: protein_or_peptide / ID: 3 / Details: GWHPBJSH015007 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 58.296727 KDa |
| Sequence | String: MSFRTRASSW TRLLLRRNPR SSFSRQISSL DASGSAFPGR ITPWISSQTH RFSSATTISP AEADRVVRDL LAEVEKEKQR EREERQRQG LDCKDIDDED EDEEDYLGIE PFIEKLKKQN LKDDGELNRR EESSDSDSEL DEVDWDEERK KEDMFNKKFQ R HKELLQTL ...String: MSFRTRASSW TRLLLRRNPR SSFSRQISSL DASGSAFPGR ITPWISSQTH RFSSATTISP AEADRVVRDL LAEVEKEKQR EREERQRQG LDCKDIDDED EDEEDYLGIE PFIEKLKKQN LKDDGELNRR EESSDSDSEL DEVDWDEERK KEDMFNKKFQ R HKELLQTL TKSETLDEAY KWMTKLDKFE EKHFKLAPEY RVIGELMNRL KVAEGKDKFI LQQKINRAMR LVEWKEAFDP NN PANYGVI ERDNVQGGEE KEERLVADGS KDDNDDNEEE FDDMKERDDI LLEKLNAIDK KLESKLSELD HTFGKKGKRL EEE IRDLAE ERNALTEKKR QPLYRKGYDV HVIDVKKVAK VTKGGRVERY TALMVCGNYE GVIGYAKAKA ETGQSAMQKA YEKC FQNLH YIERHEEHTI AHAIQTSYKK TKLYLWPAPT TTGMKAGRVV KTMLLLAGFK NIKSKVIGSR NSYNTVKAVL KALNA VETP KDVQEKFGRT VVEKYLL |
+Macromolecule #4: bS6m
| Macromolecule | Name: bS6m / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.011447 KDa |
| Sequence | String: MPLYDCMLLF KPIIRKEGLI DLVARIGKHV YSKNGVLTEV KSFGKVELGY GIKKLDGRHY QGQLMQITMM ATPNINKELH YLNKDDKLL RWLLVKHRDI KIGSSEMEDR QDDSNRVTNR SLYDESSTDE DDDILGLSR |
+Macromolecule #5: uS7m,Small ribosomal subunit protein uS7m
| Macromolecule | Name: uS7m,Small ribosomal subunit protein uS7m / type: protein_or_peptide / ID: 5 Details: GWHPBJSH000704 Extension in N-ter not present in UNIPROT,Extension in N-ter not present in UNIPROT,GWHPBJSH000704 Extension in N-ter not present in UNIPROT,Extension in N-ter not present in UNIPROT Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.955059 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) GGLDGEQKLL IKKLVNFRMK EGKRTRVRAI VYQTFHRPA RTERDVIKLM VDAVENIKPI CEVAKVGVAG TIYDVPGIVA RDRQQTLAIR WILEAAFKRR ISYRISLEKC S FAEILDAY ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) GGLDGEQKLL IKKLVNFRMK EGKRTRVRAI VYQTFHRPA RTERDVIKLM VDAVENIKPI CEVAKVGVAG TIYDVPGIVA RDRQQTLAIR WILEAAFKRR ISYRISLEKC S FAEILDAY QKRGSARRKR ENLHGLASTN RSFAHFRWW UniProtKB: Small ribosomal subunit protein uS7m |
+Macromolecule #6: uS8m
| Macromolecule | Name: uS8m / type: protein_or_peptide / ID: 6 / Details: GWHPBJSH002412 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.874127 KDa |
| Sequence | String: MGRRILNDAL RTIVNAERRG KASVELKPIS TVMSSFLRIM KEKGYIKNFQ VHDPHRVGRI TVDLQGRVND CKALTYRQDV KAKEIEEYT ERTLPTRQWG YVVVTTPDGI LDHEEAIKRN VGGQVLGFFY |
+Macromolecule #7: uS9m
| Macromolecule | Name: uS9m / type: protein_or_peptide / ID: 7 / Details: GWHPBJSH080139 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.32841 KDa |
| Sequence | String: MLSRLFLRSS NLRLATLVSS KSNSQIFSSF IRPLSTNSSG GGGGNDSGNG GNRNDAPWSF SGVNDGKSDP FSSFESGSPG GDGKWPKEE PKRWNMKEEG DEKAVFGDVS NGFGDVKSSG WDVSSKPWDL KEDEEDGKVV FDTSGEMPVS FDNSLVNEEE E RAKKELFE ...String: MLSRLFLRSS NLRLATLVSS KSNSQIFSSF IRPLSTNSSG GGGGNDSGNG GNRNDAPWSF SGVNDGKSDP FSSFESGSPG GDGKWPKEE PKRWNMKEEG DEKAVFGDVS NGFGDVKSSG WDVSSKPWDL KEDEEDGKVV FDTSGEMPVS FDNSLVNEEE E RAKKELFE KEEKELTEVI KGPDRAFGDL IAKSGITDEM LDSLIALKDF QGVQGLPPLT EIENLRREKS SKKSSRAEIE LQ MQEEIAK ARVRQVDEAG RAYGTGRRKC SIARVWIVPG KGKFLVNDKE FDVYFPMLDH RAALLRPLAE TKTLGSWDIN CTV TGGGTT GQVGAIQLGI SRALQNWEPD MRTALRAAGF LTRDSRVVER KKPGKAKARK SFQWVKR |
+Macromolecule #8: uS10m
| Macromolecule | Name: uS10m / type: protein_or_peptide / ID: 8 / Details: GWHPBJSH050210 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.251248 KDa |
| Sequence | String: MIGVLRRSSL PSRHALSAAL TSFDSGLSLN ITSAATGASV KLNLVGNGST LSSASFVISA RDFHIRSEPS MVSPAKISFR GYATGSESS IGDSAKIKIS PENVKPLSKK KKGGGESTSK IKGTRICIAI RSFDNPEKEA WCLPPHSRYA AMPDTRTLYT V LRSPHVDK ...String: MIGVLRRSSL PSRHALSAAL TSFDSGLSLN ITSAATGASV KLNLVGNGST LSSASFVISA RDFHIRSEPS MVSPAKISFR GYATGSESS IGDSAKIKIS PENVKPLSKK KKGGGESTSK IKGTRICIAI RSFDNPEKEA WCLPPHSRYA AMPDTRTLYT V LRSPHVDK KSREQFEMRF KKRFLVIKAQ SHELSNKLFW LKRYRILGAQ YELQFHCKTR LDMSPVLGSK |
+Macromolecule #9: uS11m
| Macromolecule | Name: uS11m / type: protein_or_peptide / ID: 9 / Details: GWHPBJSH051931 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.108223 KDa |
| Sequence | String: MNGVSRHLRA SSLPSLIRSY GGIKSVCRFS CQSDGSSGGR FGEQGRVPGD SENKSGLPDP NASPSKSFSE MRSGLLNKIG DGGYSAPNA PPTFRNPLRS RLPNTLPSQS GQMNQEGLPN TGGTGFSAPS YQSFTQSNLL KDNFRTGGIS KDLDFVRGVI E DDEGRRAT ...String: MNGVSRHLRA SSLPSLIRSY GGIKSVCRFS CQSDGSSGGR FGEQGRVPGD SENKSGLPDP NASPSKSFSE MRSGLLNKIG DGGYSAPNA PPTFRNPLRS RLPNTLPSQS GQMNQEGLPN TGGTGFSAPS YQSFTQSNLL KDNFRTGGIS KDLDFVRGVI E DDEGRRAT GLFSHFHRPN LETNADIIHI KLLRNNTFVT VTDSKGNVKC KATSGSLPDL KGGRKMTSYT ADATAENIGR RV KAMGLKS VVVKVNGFTH FKKKRNAIVA FRSGYSNSRN DQNPIVYIED TTRKAHNGCR LPRKRRV |
+Macromolecule #10: uS12m
| Macromolecule | Name: uS12m / type: protein_or_peptide / ID: 10 / Details: GWHPDUBS019876 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.283795 KDa |
| Sequence | String: MPTFNQLIRH GREEKRRTDR TRALDKCPQK TGVCLRVSTR TPKKPNSALR KIAKVRLSNR HDIFAYIPGE GHNLQEHSTV LIRGGRVKD LPGVKFHCIR GVKDLMGIPG RRRGRSKYGA EKPKSI |
+Macromolecule #11: uS13m
| Macromolecule | Name: uS13m / type: protein_or_peptide / ID: 11 / Details: GWHPBJSH064799 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.398301 KDa |
| Sequence | String: MLGLRRSAAT LFDHSQSLLR NLSFHGLRVQ GIRVGNAEVP NHKPLKTGLQ EVYGIGRRKS HQVLCGLGIT NKLARDLTGK ELIDLREEV GMHQHGDELR RRVGSEIQRL VEVDCYRGSR HRHGMPCRGQ RTKTNARTKK GKRVAIAGKK KAPRK |
+Macromolecule #12: uS14m
| Macromolecule | Name: uS14m / type: protein_or_peptide / ID: 12 / Details: GWHPDUBS019475 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.92885 KDa |
| Sequence | String: MANLRGVLTN VSSYCQRSFS QSALLWRSAV KPPNNLAQIQ SRQISTSLAK HSGTEQGVKR NSADHRRRLL AARFELRRKL YKAFCKDPE LPSEMREKNR YKLSKLPRNS AFTRIRNRCV FTGRSRSVTE LFRMSRICFR GLANKGELVG IKKSSW |
+Macromolecule #13: uS15m
| Macromolecule | Name: uS15m / type: protein_or_peptide / ID: 13 / Details: GWHPBJSH065138 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.211242 KDa |
| Sequence | String: MAFHLARRKQ RLHSNPYLIH LFSTSSSSAS PQDGNESGEQ QPSQSPSDFK ISSYFSGIKS SLKQQPQHQA QDGRRQLAGF DPKAPSFSG SGGDFQDVRR NLNEFRRRAA APPARDLQDL YKQNVLSKSG DLGAGKVEGS PFENLKKNLR QMRPQETRWS N LSSLQSIM ...String: MAFHLARRKQ RLHSNPYLIH LFSTSSSSAS PQDGNESGEQ QPSQSPSDFK ISSYFSGIKS SLKQQPQHQA QDGRRQLAGF DPKAPSFSG SGGDFQDVRR NLNEFRRRAA APPARDLQDL YKQNVLSKSG DLGAGKVEGS PFENLKKNLR QMRPQETRWS N LSSLQSIM KAKDSGNVRS NVFGGGEGLP ISVFGEEIEE RKKMGDDSEE MKSEFIKSYN TKELGEILRQ YRPEGKREEG WF SLQELNQ RLVKLREVEE AAAQGARKGI AFDDLRSEIQ QARKSQAFQN LDFFSVLNGT PKYLLEPPKD ELVQTYFHPD NMS SAEKMK IELAKVREEF KMSESDCGSA RVQVAQLTTK IKHLSSVLHK KDKHSKKGLI AMVHRRKKLL KYMRRTDWDS YCLS LSKLG LRDNPDYKF |
+Macromolecule #14: bS16m
| Macromolecule | Name: bS16m / type: protein_or_peptide / ID: 14 / Details: GWHPBJSH018119 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.330757 KDa |
| Sequence | String: MVVRLRLSRL GCKNRPFFRV MATDSRSPRD GKHIEVLGYF NPLPGQDGGK RMGLKFDRIK YWLSVGAQAS DPVQRLLFRS GLLPPPPMV AMGRKGGERD TRPVDPMTGR FVDAEKKTPV IIDNQPKEED NAQDKSP |
+Macromolecule #15: uS17m
| Macromolecule | Name: uS17m / type: protein_or_peptide / ID: 15 / Details: GWHPBJSH049400 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.090024 KDa |
| Sequence | String: MKPVIGTVVS NKMQKSVVVA VDRLFHNKLY NRYVKRTSKF MAHDETNACN IGDRVKLDPS RPLSKHKNWI VAEIIKKARI YSPQAAAAA ALSFSAAKSP SESSVPPVSS S |
+Macromolecule #16: bS18m
| Macromolecule | Name: bS18m / type: protein_or_peptide / ID: 16 / Details: GWHPBJSH076274 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.106117 KDa |
| Sequence | String: MKLAHSAIRS LSDVASFQTC RPFVSRSLST NANQDDWMFG GKDEKANSFF QHLGKAEKDK RDYTGFSRSQ GNGSRFDPSS DGVDGKLKE AALIYNVDDD EGVKDGYSFR PDVNSWGVNQ FPRDMNFRRQ MQRPRQNNKA EITTEEVLKK ADFRNVRFLA Q FITEAGIL ...String: MKLAHSAIRS LSDVASFQTC RPFVSRSLST NANQDDWMFG GKDEKANSFF QHLGKAEKDK RDYTGFSRSQ GNGSRFDPSS DGVDGKLKE AALIYNVDDD EGVKDGYSFR PDVNSWGVNQ FPRDMNFRRQ MQRPRQNNKA EITTEEVLKK ADFRNVRFLA Q FITEAGIL VKRKQTGISA KAQRKIAREI KTARAFGLMP FTTMGTKAFT FGKTMENRDQ DFEYEVIDDD DEYDDNTPE |
+Macromolecule #17: uS19m
| Macromolecule | Name: uS19m / type: protein_or_peptide / ID: 17 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.675223 KDa |
| Sequence | String: MAFCTKLGGH WKQGVNVPVS SMLGSLRYMS TKLYIGGLSP GTDEHSLKDA FSSFNGVTEA RVMTNKVTGR SRGYGFVNFI SEDSANSAI SAMNGQELNG FNISVNVAKD WPSLPLSLDE SIEEAEKKEN KMMSRSVWKD PFVDAFLMKK KNAALNRKIW S RRSTILPE ...String: MAFCTKLGGH WKQGVNVPVS SMLGSLRYMS TKLYIGGLSP GTDEHSLKDA FSSFNGVTEA RVMTNKVTGR SRGYGFVNFI SEDSANSAI SAMNGQELNG FNISVNVAKD WPSLPLSLDE SIEEAEKKEN KMMSRSVWKD PFVDAFLMKK KNAALNRKIW S RRSTILPE YVDSAVRIYN GKTHVRCKIT EGKVGHKFGE FAFTRKVKKH AKAK |
+Macromolecule #18: bS21m
| Macromolecule | Name: bS21m / type: protein_or_peptide / ID: 18 / Details: GWHPBJSH012122 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.541774 KDa |
| Sequence | String: MNTIAKRVTG LLTRPSHSQL QQERGIRVKV FSGDLDKALT ILQRKMQSSG MERLIKAQQT HHIKNSEKKV LARKNLERKI KSIDFARKL QSILIKKVRG L |
+Macromolecule #19: bTHXm
| Macromolecule | Name: bTHXm / type: protein_or_peptide / ID: 19 / Details: GWHPDUBS022287 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.582672 KDa |
| Sequence | String: MAAAMQWCSS MTRRIMMMQR TSPALACSAR YSSLCPAAAS MEVCGRGDSK TKKGKRFKGS YGNSRGKKQK MIERIKDKLE LPRSTPWPL PFKLI |
+Macromolecule #20: mS23
| Macromolecule | Name: mS23 / type: protein_or_peptide / ID: 20 / Details: GWHPBJSH071753 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.90348 KDa |
| Sequence | String: MSWMKGDLLS KTRRLVGGLA TREPVWLKAM EASPPPVFPR SNGNLKKIVL PEDPYVRRFA RKHPEAKLVE PNKVSAFIPD PARVYGCRV LELTKNGISE DDAMSVANME YLAERKEKKK AYKRLKELAV LQDKAPPPKP YLSTKKEMQT QEKKPPTDPP S VRRLVNQL ...String: MSWMKGDLLS KTRRLVGGLA TREPVWLKAM EASPPPVFPR SNGNLKKIVL PEDPYVRRFA RKHPEAKLVE PNKVSAFIPD PARVYGCRV LELTKNGISE DDAMSVANME YLAERKEKKK AYKRLKELAV LQDKAPPPKP YLSTKKEMQT QEKKPPTDPP S VRRLVNQL KQQKDVLLQD KPGGSANQDH WIDE |
+Macromolecule #21: mS26
| Macromolecule | Name: mS26 / type: protein_or_peptide / ID: 21 / Details: GWHPBJSH068774 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.093625 KDa |
| Sequence | String: MSLCNQLSRL VKQSPLTKQS LRSFSAEAAT HHRHHQETHS FLEPESYIGS WEAPKDPKDA ERMLAQLRRD YAKKVSLYRK DYVHEIEML RVEKQRKDEA RLVAERAANE ERRRLKAEAA KVRAEERKIF QDEFRKTLMK ERAEKLEFWR MTGLKREEKK K ARKKLLHE ...String: MSLCNQLSRL VKQSPLTKQS LRSFSAEAAT HHRHHQETHS FLEPESYIGS WEAPKDPKDA ERMLAQLRRD YAKKVSLYRK DYVHEIEML RVEKQRKDEA RLVAERAANE ERRRLKAEAA KVRAEERKIF QDEFRKTLMK ERAEKLEFWR MTGLKREEKK K ARKKLLHE QSSLWIEPKE LEKKITEALV DSTTL |
+Macromolecule #22: mS29
| Macromolecule | Name: mS29 / type: protein_or_peptide / ID: 22 / Details: GWHPBJSH054439 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 54.288594 KDa |
| Sequence | String: MFRSVVRITS RNFNLNRRSL IPNDAVSHFP SPPSASAATL IPSLDHAHRR RFSSSKSTKS SITKTKKSEG KKSKSKGGES GAAAGAEDG EFGGSAGDDL EAGRAKRLAD DEKTPSLDVG PNGRPLFTPR DVTLSKLSQK DIGSYFNFDE AALKAVLPEG L ASGIEDEF ...String: MFRSVVRITS RNFNLNRRSL IPNDAVSHFP SPPSASAATL IPSLDHAHRR RFSSSKSTKS SITKTKKSEG KKSKSKGGES GAAAGAEDG EFGGSAGDDL EAGRAKRLAD DEKTPSLDVG PNGRPLFTPR DVTLSKLSQK DIGSYFNFDE AALKAVLPEG L ASGIEDEF KESWRPALLV RKSFLDLRDN FRRIADPPMW PSEGKGGVKP KKQIILDGPV KSGKSIALAM LVHWARDEGW LV LYAPKGR DWTHGGYFYK NQHTGFWDTP LQAESILKDF VKFNEPRLRE LRCNVYDPIV LGEGAGVGYL KGQETMPIPE DST LYDLVQ MGINSTHAAV SVVVRLRKEL SLVKDVPVLI AIDQYNNWFT FSEFEEPVTP RSCRPIHARE LTTVNAFRSM MHDD MMVGA FSHSTAVGKL RKDLPDVPAD ARQNFPRYSL DEAEAVCYYY LRQRLVRREV FSEENWKKIY YLANGNGAEM RWLVP FMR |
+Macromolecule #23: mS31/mS46
| Macromolecule | Name: mS31/mS46 / type: protein_or_peptide / ID: 23 / Details: GWHPBJSH057126 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.476242 KDa |
| Sequence | String: MRGAIGRRLS NPSNGFSLAS IVKHTPFLVQ PTSHFSSSSS DGGAVRGRGR GRGGSGSPAA GAGQFGFNRE PEKANEPVGQ ARGSSESQS PGGYGHGRGR PIQSDPISPP FSSFVKPDSH SVGRGRGSLR SDPVSPFSPE PPRHPAPPQP QQPRFEPAKD G VQGSPPFA ...String: MRGAIGRRLS NPSNGFSLAS IVKHTPFLVQ PTSHFSSSSS DGGAVRGRGR GRGGSGSPAA GAGQFGFNRE PEKANEPVGQ ARGSSESQS PGGYGHGRGR PIQSDPISPP FSSFVKPDSH SVGRGRGSLR SDPVSPFSPE PPRHPAPPQP QQPRFEPAKD G VQGSPPFA KLEATKDATS PPPPPPPGAP NNISNALGSG AGRGKPFVQN EDNRHIQRSP QPLQRQKRVQ PPKDTTPRPQ LS PEEAGRR ARSQLSRGEA EGGSGGGVRG RGGGRGRGRG ARGRGRGRGG EGWRDDKKEE EAEQEALSVF VGDNADGEKF AKK MGDEIM AKLAEGYEDI CERALPSTAH DALVDAYDTN LMIECEPEYL MPDFGSNPDI DEKPPMPLRE CLEKVKPFIV ALEG IKDQE EWEEAIGEAM AEAPRMKEIV DHYSGPDRVT AKKQNEELDR IATTVPQSAP DSVKRFADRV ALSLKSNPGW GFDKK YQFM DKLVLEVSQN YK |
+Macromolecule #24: mS33
| Macromolecule | Name: mS33 / type: protein_or_peptide / ID: 24 / Details: GWHPDUBS036572 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.631559 KDa |
| Sequence | String: MASGSLKNLV TSAVTVGVTE ARARIFGHML NPTGQRSPHK ILRKKLFGDK VAEWYPYDIK NEDPNVLARE EKERLSKLEM LKRRNKGPP QKGHGRRAAK RNK |
+Macromolecule #25: mS34
| Macromolecule | Name: mS34 / type: protein_or_peptide / ID: 25 / Details: GWHPBJSH018393 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.650186 KDa |
| Sequence | String: MAATLMNRAI SRTETVGAFR LSLNLLRNFS APAAAAAASP STENPSSDSI KPKRRKKKNL IEVAQFLPNW GIGYHMAKAH WNGVSYEIT KINLYKDGRH GKAWGIVHKD GLRAAEAPKK ISGVHKRCWK YIPNLSKTTP ATTGSAPAAE VQAA |
+Macromolecule #28: uL2m C-ter
| Macromolecule | Name: uL2m C-ter / type: protein_or_peptide / ID: 28 / Details: GWHPBJSH030998 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.561801 KDa |
| Sequence | String: MSALVALCRA RTAASSSSSS LLNSLVRPAF RCLSTGFGDV QNKTLVAEME EKMLHMDINS MIGSSMPLGM MRIGTIIHNI EMNPGQGAK LVRAAGTNAK ILKEPASGKC LIKLPSGDTR WINARCRATI GTVSNPSHGV KKLYKAGQSR WLGIRPKVRG V AMNPCDHP ...String: MSALVALCRA RTAASSSSSS LLNSLVRPAF RCLSTGFGDV QNKTLVAEME EKMLHMDINS MIGSSMPLGM MRIGTIIHNI EMNPGQGAK LVRAAGTNAK ILKEPASGKC LIKLPSGDTR WINARCRATI GTVSNPSHGV KKLYKAGQSR WLGIRPKVRG V AMNPCDHP HGGGEGKSKS SGSRGRTSVS PWGKPCKGGY KSASVKKKKK RLAAREAKMK KK |
+Macromolecule #29: Large ribosomal subunit protein uL2mz, N-terminal part
| Macromolecule | Name: Large ribosomal subunit protein uL2mz, N-terminal part type: protein_or_peptide / ID: 29 / Details: GWHPBJSH056804 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.903914 KDa |
| Sequence | String: MRPGRARALR QFTLSTGKSA GRNSSGRITV FHRGGGSKRL LRRIDLKRST SSMGIVESIE YDPNRSSQIA LVRWIKGGCQ KKMNTIKKL APPRKILEPT TNTISGLFSF SFLPGKVDKR KVACFSPGLM AAYVVVGLPT GMPPLSSYKS AFASKSAGST K TLVKDVFF ...String: MRPGRARALR QFTLSTGKSA GRNSSGRITV FHRGGGSKRL LRRIDLKRST SSMGIVESIE YDPNRSSQIA LVRWIKGGCQ KKMNTIKKL APPRKILEPT TNTISGLFSF SFLPGKVDKR KVACFSPGLM AAYVVVGLPT GMPPLSSYKS AFASKSAGST K TLVKDVFF SAFSSPKAKR ETASLAFASS FDFPRIPVAG VSTAFFAPRM RQKVRGKSTF SLYEVQKWRT HSILWAHRIN GK AGLSWQS FRRQDTLGLV GAAGHKKSKP KTDQGNLPAK PIGERAKQLK ALRGLRAKDG ACKVDRAPVT YIIASHQLEA GKM VMNCDW S UniProtKB: Large ribosomal subunit protein uL2mz, N-terminal part |
+Macromolecule #30: uL3m
| Macromolecule | Name: uL3m / type: protein_or_peptide / ID: 30 / Details: GWHPBJSH008303 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.071527 KDa |
| Sequence | String: MAAVPRGLIS RFTQLLSIRS TTPSPSQCSF YLLRRFSSDA ELLTATGSDI TEAPTRIIDA KPGEMSPGSK RTGIIAVKCG MTALWDKWG ARVPVSILWV DDNIVSQVKT IEKEGIFALQ IGCGHKKPKH LTMPELGHFR AQGVPLKRKL REFPVTEDAL L PVGTELGV ...String: MAAVPRGLIS RFTQLLSIRS TTPSPSQCSF YLLRRFSSDA ELLTATGSDI TEAPTRIIDA KPGEMSPGSK RTGIIAVKCG MTALWDKWG ARVPVSILWV DDNIVSQVKT IEKEGIFALQ IGCGHKKPKH LTMPELGHFR AQGVPLKRKL REFPVTEDAL L PVGTELGV RHFVPGQYVD VTGITRGKGF QGVMKRWKFK GGPASHGCSK AHRKGGSTGQ RDDPGKVFKG RKMPGHMGAA QR TVKNVWV YKIDPARNLI WVRGQVPGAE GNFVFINDAC YKKPDISKLP FPTYLAPEDE DPSELEPLVA DLGEVDPFML AE |
+Macromolecule #31: uL4m
| Macromolecule | Name: uL4m / type: protein_or_peptide / ID: 31 / Details: GWHPBJSH075117 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.050207 KDa |
| Sequence | String: MAASLSKRVL RTFGSFTALG RSNATNASGN GLSSRYSDLK CGMGLLSNRK LSTSILTPDD SFPHDLLSQK TVITPDRTIG QYQDLVIPV TNFQNEDKGF MVLAGDVFDV PVRKDIIHHV VRWQLAKRQQ GTHSTKTISE VSGTGRKPWN QKGTGRARHG T LRGPQFRG ...String: MAASLSKRVL RTFGSFTALG RSNATNASGN GLSSRYSDLK CGMGLLSNRK LSTSILTPDD SFPHDLLSQK TVITPDRTIG QYQDLVIPV TNFQNEDKGF MVLAGDVFDV PVRKDIIHHV VRWQLAKRQQ GTHSTKTISE VSGTGRKPWN QKGTGRARHG T LRGPQFRG GCVMHGPRPR SHAIKMNKQV RRLGLKIALT ARAAEGKLLV FDDLALPTHK TKNIVNYYNQ METTKKVLVV EG GPIDEKL KLATQNLHYV NILPSIGLNV YSILLHDTLV MSRDAVNKIV ERMHTPINRK |
+Macromolecule #32: uL5m
| Macromolecule | Name: uL5m / type: protein_or_peptide / ID: 32 / Details: GWHPBJSH000742 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.258312 KDa |
| Sequence | String: MFPLNFHYED VLRQDLLLKL NYANVMEVPG LCEIRVVPKA PYNFIIKNGK LAMEIPCGQK FIQTQRGSTG KSFRSNPFLG SNKDKGYVS DLARQSTLRG HGMSNFLVRI LTVMSLLDFP VEIRKNSIQF SMETEFCEFS PELEDHFEIF EHIRGFNVTI I TSANTQDE TLLLWSGFLQ KDEGETQ |
+Macromolecule #33: uL6m
| Macromolecule | Name: uL6m / type: protein_or_peptide / ID: 33 / Details: GWHPBJSH074602 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.570778 KDa |
| Sequence | String: MEAKFFRFLK IVGVGYKARA EEAGRFLYLK LGYSHEVELA VPPAVRVFCF KNNVVCCTGI DKDRVHQFAA TVRSCKPPEV YKGKGIMYV DEVVKKKVGK KSK |
+Macromolecule #34: bL9m
| Macromolecule | Name: bL9m / type: protein_or_peptide / ID: 34 / Details: GWHPDUBS006976 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.074322 KDa |
| Sequence | String: MAHVAQSRNV IRHVVSRGTA FHKTENAIHH PLLFACQGVR YRKLEVILTT GIEKLGKGGE TVKVAPGYFR NHLMPKLLAV PNIDKYAHL IREQRKMRNY EEVKEEVKHK SSEVQTKEFE KAAKRLANAN LVLRKLIDKE KFKNRSSKED KPDVQTPVTK E EIVSEVAR ...String: MAHVAQSRNV IRHVVSRGTA FHKTENAIHH PLLFACQGVR YRKLEVILTT GIEKLGKGGE TVKVAPGYFR NHLMPKLLAV PNIDKYAHL IREQRKMRNY EEVKEEVKHK SSEVQTKEFE KAAKRLANAN LVLRKLIDKE KFKNRSSKED KPDVQTPVTK E EIVSEVAR QLCVKIDPDN VVLTEPLATF GEYEVPLKFP KTIPLPPGTV QWILKVKVRG H |
+Macromolecule #35: uL10m
| Macromolecule | Name: uL10m / type: protein_or_peptide / ID: 35 / Details: GWHPDUBS004443 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.917938 KDa |
| Sequence | String: MSSLRRALLV AGKTKINLQN CHFITGINYN GLTVKQLQEL RGILRENSNT KLLVAKNTLV FKALEGTKWE SLKPCMKGMN AWIFVQTED IPAALKTFVS FQKEKKLFDN NLGGAVFEKK LYGPQDYKVI ETMPSRADVY GVMFGALHWP GLDLVNTLQA P SASENESA AA |
+Macromolecule #36: uL11m
| Macromolecule | Name: uL11m / type: protein_or_peptide / ID: 36 / Details: GWHPBJSH065595 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.034994 KDa |
| Sequence | String: MAAVAKDIVT RRAVAGTIKL TVLAGKASPG PPVSPALGPY RLNMMAFCKD FNARTQKYKP DTPMAVKITA YMDHSFEFSV KSPTVSWYI KKAAGVDKGS TRPGHLTVTT LSVRHVYEIA KVKQTDPFCQ YMPLESICKS IIGTANSMGI KIVQDLE |
+Macromolecule #37: uL13m
| Macromolecule | Name: uL13m / type: protein_or_peptide / ID: 37 / Details: GWHPBJSH029441 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.166916 KDa |
| Sequence | String: MATQAAAASF NGNLKKAVAG IKRINLDGLR WRVFDAKGQV LGRLASQIST VLQAKDKPTY CPNRDDGDIC IVLNAKDIGI TGRKLTDKF YRWHTGYVGH LKERSLKDQM AKDPTEVIRK AVWRMLPTNN LRDDRDRKLR IFEGNEHPFG DKPLEPFVMP P RTVREMRP ...String: MATQAAAASF NGNLKKAVAG IKRINLDGLR WRVFDAKGQV LGRLASQIST VLQAKDKPTY CPNRDDGDIC IVLNAKDIGI TGRKLTDKF YRWHTGYVGH LKERSLKDQM AKDPTEVIRK AVWRMLPTNN LRDDRDRKLR IFEGNEHPFG DKPLEPFVMP P RTVREMRP RARRAMIRAQ KKAELAENGG TEVKKGKKRT PSQVTA |
+Macromolecule #38: uL14m
| Macromolecule | Name: uL14m / type: protein_or_peptide / ID: 38 / Details: GWHPBJSH089764 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.926344 KDa |
| Sequence | String: MAAALASRIS RGGRSLLGGL KNDFAGSMTS STGMMNGSIL LPQQQQRRTF IQMGTVLKVV DNSGAKKVMC IQALKGKKGA RLGDTIVAS VKEAMPNGKV KKGAVVYGVV VRAAMQRGRV DGSEVRFDDN AVVLVDNKDK KTKTDRQPIG TRVFGPVPHE L RKKKHLKI LALAQHIA |
+Macromolecule #39: uL15m
| Macromolecule | Name: uL15m / type: protein_or_peptide / ID: 39 / Details: GWHPBJSH025994 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.222631 KDa |
| Sequence | String: MIRRRISSII STSLFNSSIH HSAKPRFIVS SPLLQCRRSP ILTQVSSFPG GVSSFQGIRA YSLLSLNDLR DNVPRKLKTR KGRGIGSGK GKTAGRGHKG QKARGTMKFG FEGGQTPLRR RLPKRGFKNK FKLHFQPVGL GKIAKLINAG QIDSHELITM K TLKDVGAI ...String: MIRRRISSII STSLFNSSIH HSAKPRFIVS SPLLQCRRSP ILTQVSSFPG GVSSFQGIRA YSLLSLNDLR DNVPRKLKTR KGRGIGSGK GKTAGRGHKG QKARGTMKFG FEGGQTPLRR RLPKRGFKNK FKLHFQPVGL GKIAKLINAG QIDSHELITM K TLKDVGAI GKQIEDGVRL MGRGADEIKW PLHFEVSRVT VRAKEVVEAA GGSVRRVYYN KLGLRALLKP EWFEKKGRLL PK AARPPPK QLDRVDSIGR LPAPKKPIPF YAAEESKVES PVES |
+Macromolecule #40: uL16m
| Macromolecule | Name: uL16m / type: protein_or_peptide / ID: 40 / Details: GWHPBJSH064047 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.186928 KDa |
| Sequence | String: MYLTIKSIML LWKYLLVTES QVSKCGFHIV KKKGDVLYPK RTKYSKYRKG RCSRGCKPDG TKLGFGRYGI KSCKAGRLSY RAIEAARRA IIGHFHRAMS GQFRRNGKIW VRVFADLPIT GKPTEVRMGR GKGNPTGWIA RVSTGQILFE MDGVSLANAR Q AATLAAHK LCLSTKFVQW S |
+Macromolecule #41: bL17m
| Macromolecule | Name: bL17m / type: protein_or_peptide / ID: 41 / Details: GWHPBJSH087064 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.612463 KDa |
| Sequence | String: MTKLRKLGRH TGHRMSMLRT LVSQLVKHER IETTITKAKE VRRLADNMVQ LGKEGSLDAA RRAAGFVRGD DVIHKIFTEF AYRYKDRTG GYTRMLRTRI RVGDAAPMAY IEFIDRENEL RQSNPPTPQP PQRVPLDPWA RSRLMRQYAP PKEEKNSDSD L |
+Macromolecule #42: uL18m
| Macromolecule | Name: uL18m / type: protein_or_peptide / ID: 42 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.654122 KDa |
| Sequence | String: MVIPPAVRPP RLFDYLKPYV LKMHFTNKFV HAQVIHSPTA TVACAASSQE KALRETMGVT RDVAAASKIG KILGERLLMK DIPAVAIHM KKEQRYHGKV KAVIDSVREA GVKLL |
+Macromolecule #43: bL19m
| Macromolecule | Name: bL19m / type: protein_or_peptide / ID: 43 / Details: GWHPBJSH034513 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.348527 KDa |
| Sequence | String: MQSIRSSTRL LHRNRFSNAN TLPRFTSSSS PTLPANESRS VRGFDSSRST GAFDSPAKYL ISSLTKAVSF SSALPKHNLT YSSRCFSTV GDSVQSAPQG FPVSAPDAPP RIKFKRLDKT AKHIMQIVDK EAVEEVRTRR EIPEIRPGYI VQLKVEVPEN K RRVSIVKG ...String: MQSIRSSTRL LHRNRFSNAN TLPRFTSSSS PTLPANESRS VRGFDSSRST GAFDSPAKYL ISSLTKAVSF SSALPKHNLT YSSRCFSTV GDSVQSAPQG FPVSAPDAPP RIKFKRLDKT AKHIMQIVDK EAVEEVRTRR EIPEIRPGYI VQLKVEVPEN K RRVSIVKG IVIARRNAGL NSTFRIRRLV AGVGVESMFP LYSPNLREIK VLDKKRVRRA KLYYLRDKMN ALKKH |
+Macromolecule #44: bL20m
| Macromolecule | Name: bL20m / type: protein_or_peptide / ID: 44 / Details: GWHPDUBS041804 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.749137 KDa |
| Sequence | String: MNKKEIFKLA KGFRGRAKNC IRIARERVEK ALQYSYRDRR NKKREMRGLW IERINAGSRQ HGVNYGNFIH GLMKENIQLN RKVLSELSM HEPYSFKALV DVSRNSFPGN KNVVQAPRKV DVTSINA |
+Macromolecule #45: bL21m
| Macromolecule | Name: bL21m / type: protein_or_peptide / ID: 45 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.955697 KDa |
| Sequence | String: MASLRCFREL SRRATTVFSI NQTRSISSFH GIEFSGTSIS HGTVIPNRSL TRNLPWYSHW YRSQDRCFSS NTKDTDEDEE SSEGEDDDE EEGEDFEDSA DMEVEREYSP AEKVEEAEEI GYKVMGPLKP SERLFKPYEP VFAIVQIGSH QFKVSNGDSI F TEKLKFCD ...String: MASLRCFREL SRRATTVFSI NQTRSISSFH GIEFSGTSIS HGTVIPNRSL TRNLPWYSHW YRSQDRCFSS NTKDTDEDEE SSEGEDDDE EEGEDFEDSA DMEVEREYSP AEKVEEAEEI GYKVMGPLKP SERLFKPYEP VFAIVQIGSH QFKVSNGDSI F TEKLKFCD INDKLELTKV LLLGSASQTI IGRPILPDAT VHAVVEEHAL DEKVLIFKKK RRKNYRRTRG HRQELTKLRI TD IQGIEKP EPKIVHKPSK EAVTEQTKAE LVA |
+Macromolecule #46: uL22m
| Macromolecule | Name: uL22m / type: protein_or_peptide / ID: 46 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.793791 KDa |
| Sequence | String: MAGWQRNLQL IRHVARRVTN SNVSTANYSW SLESPFSKGC LLRPTYSSAP LHHYLKQVGI STSRKLEAGE EPVSSPLSSP ALLGPGKEE EEQKIIPKRK KVQAVLKAIK QSPKKVNLVA ALVRGMRVED ALMQLQVTVK RASHTVYRVI HAARANASHN H GLDPDRLI ...String: MAGWQRNLQL IRHVARRVTN SNVSTANYSW SLESPFSKGC LLRPTYSSAP LHHYLKQVGI STSRKLEAGE EPVSSPLSSP ALLGPGKEE EEQKIIPKRK KVQAVLKAIK QSPKKVNLVA ALVRGMRVED ALMQLQVTVK RASHTVYRVI HAARANASHN H GLDPDRLI IAEAFVGKGL FKKRISIHGK GKCGLMIRPE CRLTVIVRET TPEEEAEIAR LKVHNFKKLT KRERRLNPHK LI ETTPIWN RRGTKANHRS SELVPSR |
+Macromolecule #47: uL23m
| Macromolecule | Name: uL23m / type: protein_or_peptide / ID: 47 / Details: GWHPBJSH001034 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.963191 KDa |
| Sequence | String: MGSRLGRRVV HFANLPIKLL MPTKLTNIHE FALRTIPSAT KIEIKRVLES LYGFEVEKVN TLNMDGKKKK RGGLLIAKAD YKKAYVTLK NPLSISGDLF PVKFIEEDRK SKVKGSSFVE EEDDKKSHWL DQKERREVGG YGNGKGRRGG GGERSASPAK G VASAAAVA KFPWSSMRFG GK |
+Macromolecule #48: uL24m
| Macromolecule | Name: uL24m / type: protein_or_peptide / ID: 48 / Details: GWHPBJSH068111 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.547445 KDa |
| Sequence | String: MGWKAAEKLI RHWKILRGDN VMIIRGKDKG ETGTVKRVIR SQNRVIVEGK NLIKKHIKGG PDHEGGIFTV EAPLHASNVQ VVDPVTGRP CKVGVKYLED GTKVRVARGT GTSGSIIPRP EILKIRTTPR PTTAGPKDTP MEFVWEQTYD AKTGKGMPDL |
+Macromolecule #49: bL25-2m
| Macromolecule | Name: bL25-2m / type: protein_or_peptide / ID: 49 / Details: GWHPBJSH070969 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.376742 KDa |
| Sequence | String: MAKWWRGLRS VTAEVADLPA SSFGRSIRCI HQYQTIQAIP REATGRGVSA RDRTIGRIPA VVFPQSLLDT DASKRGVSRK QLLTADKKQ IKSIIDSVGL PFFCSTTFQL QIRAGQGSST LVESGRVLPL KVHRDEETGK ILNLVFVWAD DGEKLKVDVP V VFKGLDHC ...String: MAKWWRGLRS VTAEVADLPA SSFGRSIRCI HQYQTIQAIP REATGRGVSA RDRTIGRIPA VVFPQSLLDT DASKRGVSRK QLLTADKKQ IKSIIDSVGL PFFCSTTFQL QIRAGQGSST LVESGRVLPL KVHRDEETGK ILNLVFVWAD DGEKLKVDVP V VFKGLDHC PGLQKGGNLR TIRSTLKLLG PAEHIPSKIE VDVSNLDIED KVLLQDVVFH PSLKLLSKNE TMPVCKIVAT SP VKEPEAV QA |
+Macromolecule #50: bL25m
| Macromolecule | Name: bL25m / type: protein_or_peptide / ID: 50 / Details: GWHPBJSH022473 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.945787 KDa |
| Sequence | String: MLLRRATALP KTLQNLRLFS PAATSALALD HTLEIQLEYL PGFPRPDAKH AETILAVPRS DSGKNISAKE RKAGRVPSII FEQEDGQHG GNKRLVSVQT NQIRKLVTHL GYSFFLSRLF DVEVRSEIGS DEVIEKVRAL PRSIHLHSGT DAPLNVTFIR A PPGTLLKV ...String: MLLRRATALP KTLQNLRLFS PAATSALALD HTLEIQLEYL PGFPRPDAKH AETILAVPRS DSGKNISAKE RKAGRVPSII FEQEDGQHG GNKRLVSVQT NQIRKLVTHL GYSFFLSRLF DVEVRSEIGS DEVIEKVRAL PRSIHLHSGT DAPLNVTFIR A PPGTLLKV DIPLVFIGDD VSPGLKKGAS LNTIKRTVKF LCPAEIVPPY IEVDLSLLDV GQKLVAGDLK VHPALKLIRP KD EPIVKIA GGRVSDQQKD QQKKDQPKKE QSKK |
+Macromolecule #51: bL27m
| Macromolecule | Name: bL27m / type: protein_or_peptide / ID: 51 / Details: GWHPBJSH034173 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.095584 KDa |
| Sequence | String: MNFFNSAASL CRRVSLRELI TEVPAYGGSG IYDGSSSGLS LVFKRWATKK TAGSTKNGRD SKPKNLGVKK FGGENVIPGN IIVRQRGTR FHPGDYVGIG KDHTLFALKE GRVRFEKNKI TGRKWIHVDP KGGHVLHPIY TRAAATKSTQ METASSS |
+Macromolecule #52: bL28m
| Macromolecule | Name: bL28m / type: protein_or_peptide / ID: 52 / Details: GWHPBJSH021367 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.112625 KDa |
| Sequence | String: MAFRGKEMMK KLVKKVGAEN ITPELKEKLK ACVPDTKVVM GRAKRGLYAG RHIQYGNRVS EDGGNKSRRC WKPNVQEKRL FSYIFDSHI KVKVTTHALR CIDKAGGIDE YLLKTPYQKM DTEMGLYWKT KVEARYAELG QMEVAFFNPE DEAKLEQGFK D LNIAKKDA ...String: MAFRGKEMMK KLVKKVGAEN ITPELKEKLK ACVPDTKVVM GRAKRGLYAG RHIQYGNRVS EDGGNKSRRC WKPNVQEKRL FSYIFDSHI KVKVTTHALR CIDKAGGIDE YLLKTPYQKM DTEMGLYWKT KVEARYAELG QMEVAFFNPE DEAKLEQGFK D LNIAKKDA RREARRTFRK KGGGNKGDEE ASIEGGGGSE SHQEDDNGWL EAKA |
+Macromolecule #53: uL29m
| Macromolecule | Name: uL29m / type: protein_or_peptide / ID: 53 / Details: GWHPDUBS024649 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.970814 KDa |
| Sequence | String: MFLTRFIGRR FLAVAASTRS ESTTAAAAST VKIAKNPLEE FFEFDRSQDE DKPVVYGRGW KASELRLKSW DDLQKLWYVL LKEKNMLMT QRQMLQAQNM MFPNPERIPK VRRSMCRIKH VLTERAIEEP DPRRSAEMKR MVNGM |
+Macromolecule #54: uL30m
| Macromolecule | Name: uL30m / type: protein_or_peptide / ID: 54 / Details: GWHPBJSH092856 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.347487 KDa |
| Sequence | String: MSGFRAFKAQ VPIQWSQSLY ITLVRGLPGT RKLHRRTLEA MGLRRCHRTV LHSNTSSIRG MIQQVKRMVV VETEEMFKAR KEAEANHKA LRPPLVVSHS IPATGSSNMS |
+Macromolecule #55: bL31m
| Macromolecule | Name: bL31m / type: protein_or_peptide / ID: 55 / Details: GWHPBJSH092851 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.785254 KDa |
| Sequence | String: MNERVFEAFV LGPNRTYWSS EAKRQARAYT FLLCITIDEG RTTPSLSKLI HLLTVAGWTM KKGVHPQMQW ISYVTQSGRL MHVMMTRIH HVGKVYHFGA KRQLAQSIGQ IAKFKRRFNE LEEEPSHENN IDNQKK |
+Macromolecule #56: bL32m
| Macromolecule | Name: bL32m / type: protein_or_peptide / ID: 56 / Details: GWHPBJSH071754 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.228754 KDa |
| Sequence | String: MALRLTSLKR AAVESCRIFQ TRSLSHVAAM PTPLNSAIDR TISMPPLISP EFDQNQNQPG SIDEKSFGFG LPDFGFGGSI ELMAVPKKK VSKHKRGIRN GPKALKPVPV IIRCRSCGRV KLPHFFCCSG ERGNPTEQGN |
+Macromolecule #57: bL33m
| Macromolecule | Name: bL33m / type: protein_or_peptide / ID: 57 / Details: GWHPDUBS049973 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.446915 KDa |
| Sequence | String: MGDKRKKTFM FIRLVSAAGT GFFYVKRKSS KGLLEKLEFR KYDPRVNRHV LFTEQKMKIV SVS |
+Macromolecule #58: bL34m
| Macromolecule | Name: bL34m / type: protein_or_peptide / ID: 58 / Details: GWHPBJSH048407 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.532242 KDa |
| Sequence | String: MAAKTLIRSG ALLMNRFLSK PTTNLVKNNL RSFQQIAPQG PELPPYFPPS LSNLQRSLYS PPNDTVTLKE LTERGFLHPS GLPSLEFFL PEVDPSSEPL LLFPKRTFQP STIRRKRNHG FFARKATKGG RRVIARRIAK GRHRVTA |
+Macromolecule #59: bL35m
| Macromolecule | Name: bL35m / type: protein_or_peptide / ID: 59 / Details: GWHPBJSH013115 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.991387 KDa |
| Sequence | String: MQRFCTKLRS ISLHSNRNIS FTSVPHRLIH HSRSPQSTLG FARTSKWNFV SPSNPTSTGS ISVPHHLVQV RNITSKDKMA KWKKKWRPR TPITSKVKKV KIKFYSSYKD RFRPLNDGTI LRWKEGKRHN AHLKSKKSKR RLRQPGLVPP AYAKVMKKLN F CN |
+Macromolecule #60: bL36m
| Macromolecule | Name: bL36m / type: protein_or_peptide / ID: 60 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.452438 KDa |
| Sequence | String: MKVRSSVKKM CEFCKTVKRR GRVYVICSSN PKHKQRQGFS SFAYEGITPS PLFAEPIASQ ELVRLPGQGV SAGLASLLHK RPMPTAYFG WRSGLASILF KQGN |
+Macromolecule #61: mL40
| Macromolecule | Name: mL40 / type: protein_or_peptide / ID: 61 / Details: GWHPDUBS047948 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.264758 KDa |
| Sequence | String: MIRITRPKPI IESLTYNLIQ RCSAGGTPKG KAKLKTGQPL KRNKLSTKNP AASGEGEEAV KGKGRISDEK QKLYEQCLNA PCPVRSLTP KEAEREAQRE KLGLISKDKQ RDMEIQKKGG AAAMGINTDE PMRIGTAGLD YVSLGIFSAD ELVKYKVTAE D GERLAKEN ...String: MIRITRPKPI IESLTYNLIQ RCSAGGTPKG KAKLKTGQPL KRNKLSTKNP AASGEGEEAV KGKGRISDEK QKLYEQCLNA PCPVRSLTP KEAEREAQRE KLGLISKDKQ RDMEIQKKGG AAAMGINTDE PMRIGTAGLD YVSLGIFSAD ELVKYKVTAE D GERLAKEN SKVLMREHRE RRAAETVLLN MKKAAIEALP EKLKMAALEP DLTPFPANRG MATLTPPIEG YLEKVMDAAK KS SSKEKLR |
+Macromolecule #62: mL41
| Macromolecule | Name: mL41 / type: protein_or_peptide / ID: 62 / Details: GWHPDUBS013782 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.075808 KDa |
| Sequence | String: MTLGLLSAIG RSFRRKRASS LDILSPKRAP RDFYKGKNCK PTGFHTKKGG YVVQPDKLPN YVIPDLTGFK LKPYVSQCPI EVNKTTEAS K |
+Macromolecule #63: mL43
| Macromolecule | Name: mL43 / type: protein_or_peptide / ID: 63 / Details: GWHPDUBS043571 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.679948 KDa |
| Sequence | String: MALRGVWQLQ KLVVSYCNWG GSSRGIRAFM ESELPALIEK NPQLEVVTEL SRGQHPYLKG IYRNRNERVV CVKNMDPDQV LLNATRLRN SLGRKVVKLR TRHVTKHPSV QGTWTTAVKF |
+Macromolecule #64: mL46
| Macromolecule | Name: mL46 / type: protein_or_peptide / ID: 64 / Details: GWHPBJSH030844 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.178295 KDa |
| Sequence | String: MPRSSLRLLA KPLLESRRGF CTSSDKIVAS VLFERLRVVI PKPDPAVYAF QEFKFNWQQQ FRRRYPDEFL DIAKNRAKGE YQMDYVPAP RITEADKNND RKSLYRALDK KLYLLIFGKP FGATSDKPVW HFPEKVYDSE PTLRKCAESA LKSVVGDLTH T YFVGNAPM ...String: MPRSSLRLLA KPLLESRRGF CTSSDKIVAS VLFERLRVVI PKPDPAVYAF QEFKFNWQQQ FRRRYPDEFL DIAKNRAKGE YQMDYVPAP RITEADKNND RKSLYRALDK KLYLLIFGKP FGATSDKPVW HFPEKVYDSE PTLRKCAESA LKSVVGDLTH T YFVGNAPM AHMAIQPTEE MPDLPSYKRF FFKCSVVAAS KYDISNCEDF VWVTKDELLE FFPEQAEFFN KMIIS |
+Macromolecule #65: mL53
| Macromolecule | Name: mL53 / type: protein_or_peptide / ID: 65 / Details: GWHPDUBS021933 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.567626 KDa |
| Sequence | String: MLKFLSKVKI EFNTLDPRLA SCVEFLAQCN ARKAKESNPN CQVLVKRRTD DQPPQISVTF VNGVEEAFDA AATSAQSIRK MILDKGQYL ETEQMFREAG EQWPVIIPEE EIHQEAPGVK PRKAEDKKQ |
+Macromolecule #66: mL54
| Macromolecule | Name: mL54 / type: protein_or_peptide / ID: 66 / Details: GWHPBJSH029436 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.767887 KDa |
| Sequence | String: MNCRIGLITG GSKDVVRRTF AAAASKAKKG GKGGGSDAPK GSSLSKEIKS TTVVGANTLK DRSDPKILPD SDYPDWLWRL LQIRPALSE LRRKNVETLP YDDLKRFVKL DTRAKIKENN SIKAKN |
+Macromolecule #67: mL59/mL64
| Macromolecule | Name: mL59/mL64 / type: protein_or_peptide / ID: 67 / Details: GWHPBJSH003404 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.415188 KDa |
| Sequence | String: MAGKAAEAVA KTVTAVQHPW RAKLDKYRTE LTKGVWGYWE MGAWKPLGIS ARRRAMLRKE VLTAGEDWPY DPERKAMRTK RKGHKCDRI SAEKRENTAK LMLKMPQMLL DYKKRRWEKK MKEEEKAKED K |
+Macromolecule #68: mL60
| Macromolecule | Name: mL60 / type: protein_or_peptide / ID: 68 / Details: GWHPDUBS017065 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.680037 KDa |
| Sequence | String: MGAFWGTRVM EIVKKHDSGG LIWKRIKLTS TRKANAKTRL RRVWQNEAVL KACGTSDAAV SPGVSSNTET CTSAVKNKE |
+Macromolecule #69: mL80
| Macromolecule | Name: mL80 / type: protein_or_peptide / ID: 69 / Details: GWHPDUBS033403 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.391539 KDa |
| Sequence | String: MWFTGGGGGV GGLRKLCRAS ATVLEKEMSC NSLLVRYMSR ERAVNVRKIN PKVSIQEAHI ISSSLYDVFK KHGPLSVPNT WLRAQEAGV SGLNSKTHMK LMLKWMRGRK MLKLICNQVG SSKKFFHTVL PDDDPQQETP PVAAAAAAAA AATTTDKNKK A FSKRRSK |
+Macromolecule #70: mL87
| Macromolecule | Name: mL87 / type: protein_or_peptide / ID: 70 / Details: GWHPBJSH067819 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.712545 KDa |
| Sequence | String: MGFGAIRSIL RPLSRTLVSR VAASCSSAPL IPAAKPELCS FLGGSRLPWI PMANHFHSLS LTDTRLPKRR PMAHPKKKRF KLKPPGPYA YVQYTPGEPI ASNNPNKGSV KRRNAKKRIG QRRAFILSEK KKRQGLVQEA KRKKRIKEVE RKMAKVARER A WEERLAEL QRLEEEKKKS MSS |
+Macromolecule #71: mL101 (rPPR4)
| Macromolecule | Name: mL101 (rPPR4) / type: protein_or_peptide / ID: 71 / Details: GWHPBJSH088794 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 55.94725 KDa |
| Sequence | String: MALRHLSRPR DIVKRSTKKY LDEPLYHRLF KDGGSEVSVR QQLNQFLKGT KHVFKWEVGD TIKKLRSRGL YYPALKLSEV MEHRGMNKT VSDQAIHLDL VAKARGIAAG ESYFVDLPET SKTELTYASL LNCYCKELMT EKAEGLLNKM KELNITVSSM S YNSLMTLY ...String: MALRHLSRPR DIVKRSTKKY LDEPLYHRLF KDGGSEVSVR QQLNQFLKGT KHVFKWEVGD TIKKLRSRGL YYPALKLSEV MEHRGMNKT VSDQAIHLDL VAKARGIAAG ESYFVDLPET SKTELTYASL LNCYCKELMT EKAEGLLNKM KELNITVSSM S YNSLMTLY TKTGQAERVP GMIQEMKAED VMPDSYTYNV WMRALAATED VSGVERVIEE MNRDGRVAPD WTTYSNMASI YV DAGLSEK AEKALQELEM KNTDRDFKAY QFLITLYGRL GKLNEVYRIW RSLRLAMPKT SNVAYLNMIQ VLVNLKDLPG AET LFKEWQ ANCSTYDIRV VNVMIGAYTR EGLVEKANEL KEKSPRRGGK LNAKTWELFM DYYVKRGETA QALECITKAV SIGK GDGGK WLPSEDTVRA LMSQFEEKKD VNGAESLLEI LKKGTDDVGA ETFESLIRTY AAAGKSHPAM RQRLKMEKVK VDKAT EKLL DELCQDE |
+Macromolecule #72: mL102 (rPPR5)
| Macromolecule | Name: mL102 (rPPR5) / type: protein_or_peptide / ID: 72 / Details: GWHPBJSH031757 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 85.828961 KDa |
| Sequence | String: MAFISRSKRY QSKARVYLSL PRSSNSSLFS LPRLFSTIEE TQTPANANPE TQSPDAKSET KKNLTSTETR PLRERFQRGK RQNHEKLED TICRMMDNRA WTTRLQNSIR DLVPEWDHSL VYNVLHGAKK LEHALQFFRW TERSGLIRHD RDTHMKMIKM L GEVSKLNH ...String: MAFISRSKRY QSKARVYLSL PRSSNSSLFS LPRLFSTIEE TQTPANANPE TQSPDAKSET KKNLTSTETR PLRERFQRGK RQNHEKLED TICRMMDNRA WTTRLQNSIR DLVPEWDHSL VYNVLHGAKK LEHALQFFRW TERSGLIRHD RDTHMKMIKM L GEVSKLNH ARCILLDMPE KGVPWDEDMF VVLIESYGKA GIVQESVKIF QKMKDLGVER TIKSYNSLFK VILRRGRYMM AK RYFNKMV SEGVEPTRHT YNLMLWGFFL SLRLETALRF FEDMKTRGIS PDDATFNTMI NGFCRFKKMD EAEKLFVEMK GNK IGPSVV SYTTMIKGYL AVDRVDDGLR IFEEMRSSGI EPNATTYSTL LPGLCDAGKM VEAKNILKNM MAKHIAPKDN SIFL KLLVS QSKAGDMAAA TEVLKAMATL NVPAEAGHYG VLIENQCKAS AYNRAIKLLD TLIEKEIILR HQDTLEMEPS AYNPI IEYL CNNGQTAKAE VLFRQLMKRG VQDQDALNNL IRGHAKEGNP DSSYEILKIM SRRGVPRESN AYELLIKSYM SKGEPG DAK TALDSMVEDG HVPDSSLFRS VIESLFEDGR VQTASRVMMI MIDKNVGIED NMDLIAKILE ALLMRGHVEE ALGRIDL LN QNGHTADLDS LLSVLSEKGK TIAALKLLDF GLERDLSLEF SSYDKVLDAL LGAGKTLNAY SVLCKIMEKG SSTDWKSS D ELIKSLNQEG NTKQADVLSR MIKKGQGIKK QNNVSL |
+Macromolecule #73: mL104 (rPPR9)
| Macromolecule | Name: mL104 (rPPR9) / type: protein_or_peptide / ID: 73 / Details: GWHPBJSH068020 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 59.234504 KDa |
| Sequence | String: MPPSLPSLQL RRLLLRSFIS SSSVNTLQSQ PRIISSKPLF SPLPPSRSSI FSTFPSRFFS SETNAESESL DSNEIALSFS KELTGNPDA ESQTISQRFN LSFSHITPNP DLILQTLNLS PEAGRAALGF NEWLDSNSNF SHTDETVSFF VDYFGRRKDF K GMLEIISK ...String: MPPSLPSLQL RRLLLRSFIS SSSVNTLQSQ PRIISSKPLF SPLPPSRSSI FSTFPSRFFS SETNAESESL DSNEIALSFS KELTGNPDA ESQTISQRFN LSFSHITPNP DLILQTLNLS PEAGRAALGF NEWLDSNSNF SHTDETVSFF VDYFGRRKDF K GMLEIISK YKGIAGGKTL ESAIDRLVRA GRPKQVTDFF EKMENDYGLK RDKESLTLVV KKLCEKGHAS IAEKMVKNTA NE IFPDENI CDLLISGWCI AEKLDEATRL AGEMSRGGFE IGTKAYNMML DCVCKLCRKK DPFKLQPEVE KVLLEMEFRG VPR NTETFN VLINNLCKIR RTEEAMTLFG RMGEWGCQPD AETYLVLIRS LYQAARIGEG DEMIDKMKSA GYGELLNKKE YYGF LKILC GIERLEHAMS VFKSMKANGC KPGIKTYDLL MGKMCANNQL TRANGLYKEA AKKGIAVSPK EYRVDPRFMK KKTKE VDSN VKKRETLPEK TARKKKRLKQ INMSFVKKPH NKMRRRM |
+Macromolecule #74: Nascent peptide
| Macromolecule | Name: Nascent peptide / type: protein_or_peptide / ID: 74 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 316.353 Da |
| Sequence | String: GVAA |
+Macromolecule #78: mS35
| Macromolecule | Name: mS35 / type: protein_or_peptide / ID: 78 / Details: GWHPBJSH049245 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.577301 KDa |
| Sequence | String: MRGALFRNAS LCARRLLLSP RINTHQPSSS SNARFVAPIA TPFPPRLFSS ESDSSGENPP PPPESSSPIE SSNKKDLAVE DVGNKELKA RIEKYFNEGN EDALPGIIEA LLQRRLADKH ADTDDEVMDS LQNQPFKDDV KDEDFESDFE EAHSTDDELE D LYNSPEYV ...String: MRGALFRNAS LCARRLLLSP RINTHQPSSS SNARFVAPIA TPFPPRLFSS ESDSSGENPP PPPESSSPIE SSNKKDLAVE DVGNKELKA RIEKYFNEGN EDALPGIIEA LLQRRLADKH ADTDDEVMDS LQNQPFKDDV KDEDFESDFE EAHSTDDELE D LYNSPEYV KKKMQNNEFF NMDEKKWDVI VREGIRHGIL KDTKECEEIL EDMLHWDKLL PDDLKKKVEA KFNELGDMCE RG EIEAEAA YELFKEFEDE MVIQYGDQME AAGPPKFDET DPSYSNTNLD DPPGKGPILR WQSRIVFAPG GDAWHPKNRK VKL SVTVKE LGLSKHQARR LRELVGKRYD SGKDELTITS ERFEHREENR KDCLRTLYGL IEEAAKANKI AEDIRTAYVK QRLQ ANPAF MQKLQAKIMR SKESNPINA |
+Macromolecule #79: mS37
| Macromolecule | Name: mS37 / type: protein_or_peptide / ID: 79 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.227926 KDa |
| Sequence | String: MGRKAGNLYI NPKKLGNLAK PCMKEMVSFL NCMALNNIKD DKCEKQKQLL SVCMQGQSDH KNKSWGNINY HLQRLTRGRK |
+Macromolecule #80: mS38
| Macromolecule | Name: mS38 / type: protein_or_peptide / ID: 80 / Details: GWHPDUBS042575 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.562171 KDa |
| Sequence | String: MANLMQRFIR NQSCLRAIPK LTPNLIPHQK PCIIINNNVE SPFPLTNPVI AFSGSGIKPD EALRFYPSFP IGYGLNPSVV HGSGLTDTV VEEKEKEVVI HADSVKKKRK KKMNKHKYRK LRKKQGRKS |
+Macromolecule #81: mS41
| Macromolecule | Name: mS41 / type: protein_or_peptide / ID: 81 / Details: GWHPBJSH068514 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.49895 KDa |
| Sequence | String: MAWMQMIQSA RFLRKTQPFS TPSLARFYSK PAPYAVKVGI PEFLSGIGGG VETHVARLET ELGDLSKLLV TRTLKLKKFG IPCKHRKLI LKYSQKYRLG LWKPRADAIK A |
+Macromolecule #82: mS45
| Macromolecule | Name: mS45 / type: protein_or_peptide / ID: 82 / Details: GWHPBJSH025481 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44.013531 KDa |
| Sequence | String: MFKLSRMIPR LQSVSLNPST TASGSLIENA PRIFQSLGNR AFSGKSSSDG SGSNENGWDI ATGGSFGGGT TGGSDDLDWD HKSMWSTGL SKEHFDGVSV GRQNPSSENA VSDSGDVMSK LGPREVAMVN EMNEYDDMIK EIEKDNRHSR VFVDGIKQKM M EMSVLLKQ ...String: MFKLSRMIPR LQSVSLNPST TASGSLIENA PRIFQSLGNR AFSGKSSSDG SGSNENGWDI ATGGSFGGGT TGGSDDLDWD HKSMWSTGL SKEHFDGVSV GRQNPSSENA VSDSGDVMSK LGPREVAMVN EMNEYDDMIK EIEKDNRHSR VFVDGIKQKM M EMSVLLKQ VKEPGARGSY LKDSEKTEMY RLHKENPEVY TVERLAKDYR IMRQRVHAIL FLKEDEEEEE RKLGRPLDDS VE RLLDEYP EFFVSHDREF HVASLSYKPD FKVMPEGWDG TIKDMDEVHY EISKKEDDIL YEEFLRRFEF NKLKWKGEVK CHK YSRRRS SDGWKITVEK LGPQGKRGNG GGWKFVSLPD GSSRPLNEVE KMYVKREAPR RRRKILP UniProtKB: Uncharacterized protein |
+Macromolecule #83: mS47
| Macromolecule | Name: mS47 / type: protein_or_peptide / ID: 83 / Details: GWHPBJSH021410 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on ester bonds; Thioester hydrolases |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.518668 KDa |
| Sequence | String: MQTVRALRRV TKPSQWVRSV SQGKRSFSAL PNFSASDADV QDQVSVEGKA KSRAAILDRP SSLNALSAPM VGRLKRLYES WEENPAISF VLMKGSGKTF CSGADVLPLY HSINEGNTEE CKHFFGSLYN FVYLQGTYLK PNIAIMDGVT MGCGGGISIP G MFRVATDK ...String: MQTVRALRRV TKPSQWVRSV SQGKRSFSAL PNFSASDADV QDQVSVEGKA KSRAAILDRP SSLNALSAPM VGRLKRLYES WEENPAISF VLMKGSGKTF CSGADVLPLY HSINEGNTEE CKHFFGSLYN FVYLQGTYLK PNIAIMDGVT MGCGGGISIP G MFRVATDK TVLAHPEVQI GFHPDAGASY YLSRLPGYLG EYLALTGQKL DGVEMIACGL ATHFCLHSRL GMVEERIGKL LT DDPTVIE ASLAQYSDLV YPDNTSVLHK IEMIDRYFGL DTVEEIIEAM ENEVADSGNE WCKKTLKQVK EASPLSLKIT LQS IREGRF QTLDQCLTRE YRISLCGVSK TVSGDFCEGI RARLVDKDFA PKWDPPRLED VSKDMVDCYF SPATDADDSE SELK LPTAQ REPYF |
+Macromolecule #84: mS83 (rPPR10)
| Macromolecule | Name: mS83 (rPPR10) / type: protein_or_peptide / ID: 84 / Details: GWHPBJSH050066 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.367781 KDa |
| Sequence | String: MQYLSNSRRI ARALKPLALL EADVRFASHV VDSTQWQNHR SPSVSVSPGM NTQSHMTGLM MLRSSFSSEA KLVEDPTEAV KELHGKILD SVNVKRSMPP NALLWSLIEN CRKEDDISFL FDALQNLRRF RLSNLRIHDN FNCNLCQQVA KTCVRVGAIN H GKRALWKH ...String: MQYLSNSRRI ARALKPLALL EADVRFASHV VDSTQWQNHR SPSVSVSPGM NTQSHMTGLM MLRSSFSSEA KLVEDPTEAV KELHGKILD SVNVKRSMPP NALLWSLIEN CRKEDDISFL FDALQNLRRF RLSNLRIHDN FNCNLCQQVA KTCVRVGAIN H GKRALWKH NVHGLTPSVA SAHHMMSYAL EHKNSNLMEE VMKLLKANDL PLQPGTADLV FRICHETDSW DLLAKYSKKF CK AGVKLRK TTFDVWMEFA AKRGDTESLW KVDKLRSETY TQHTLSAAFS CAKGFLLEHK PEEAAAVIQI ICQAYPDEKK SAL EAEKEK LVNEWPVDVL KHQNEEDKKA VAASLKSDIP AMVNALVNSG LRVSVDLDEL NKNEALLS |
+Macromolecule #85: mS77 (NFD5)
| Macromolecule | Name: mS77 (NFD5) / type: protein_or_peptide / ID: 85 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 82.64882 KDa |
| Sequence | String: MKSFLLSRQA IHRISLLSSK TPTFCRNFSA ITSTISHSDR HLRSYDEQTP FQNVEIPRPI SSFNRYFHFT RESRLSESSA AIDDSNDQE EDDEDGTTNE FLSRFVWIMR GKVSEAYPDC DKKMIDGMLL LIVEKVVEEI ERGGFNKVGS APPSPSSEFS D DLWATIWE ...String: MKSFLLSRQA IHRISLLSSK TPTFCRNFSA ITSTISHSDR HLRSYDEQTP FQNVEIPRPI SSFNRYFHFT RESRLSESSA AIDDSNDQE EDDEDGTTNE FLSRFVWIMR GKVSEAYPDC DKKMIDGMLL LIVEKVVEEI ERGGFNKVGS APPSPSSEFS D DLWATIWE VSNTVLKDME KERKKEKMKQ YVQSPEVMEM CRFAGEIGIR GDLLRELRFK WAREKMDDAE FYESLEQQRD LD NSIRESE TVDGEVEEEG FVPSDEVESR SISLPKRKGK LKYKIYGLEL SDPKWVEMAD KIHEAEEEAD WREPKPVTGK CKL VMEKLE SLQEGDDPSG LLAEWAELLE PNRVDWIALI NQLREGNTHA YLKVAEGVLD EKSFNASISD YSKLIHIHAK ENHI EDVER ILKKMSQNGI FPDILTATAL VHMYSKSGNF ERATEAFENL KSYGLRPDEK IYEAMILGYV NAGKPKLGER LMKEM QAKE LKASEEVYMA LLRAYAQMGD ANGAAGISSS MQYASDGPLS FEAYSLFVEA YGKAGQVDKA KSNFDEMRKL GHKPDD KCI ANLVRAYKGE NSLDKALRLL LQLEKDGIEI GVITYTVLVD WMANLGLIEE AEQLLVKISQ LGEAPPFELQ VSLCCMY SG VRNEKKTLQA LGVLEAKRDQ MGPNEFDKVI SALKRGGFEK DARRMYKYME ARKFLPSQRL QMDMVAPPRA FGSGSGRV R RFNS |
+Macromolecule #86: mS76 (rPPR1)
| Macromolecule | Name: mS76 (rPPR1) / type: protein_or_peptide / ID: 86 / Details: GWHPDUBS003368 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.681852 KDa |
| Sequence | String: MALLSRIRSS ASLLRHLNPS PQIRSLSSAS SILSPDSKTP LTSKQKSKTA LTLLKTERDP DRILEICRAA SLTPDCHIDR LAFSAAVQN LTENKHFSAV TNLLDGLLEN RPDLKTERFA AHAIVLYAQA NMLDHSLRVF SDLEKLEIQR TVKSLNALLF A CLVAKDYK ...String: MALLSRIRSS ASLLRHLNPS PQIRSLSSAS SILSPDSKTP LTSKQKSKTA LTLLKTERDP DRILEICRAA SLTPDCHIDR LAFSAAVQN LTENKHFSAV TNLLDGLLEN RPDLKTERFA AHAIVLYAQA NMLDHSLRVF SDLEKLEIQR TVKSLNALLF A CLVAKDYK EAKRVYIEIP KMYKIEPDLE TYDRMIKVFC ESGSASSSYS IVAEMERKGV KPTSSTFGLM IAGFYREDKK ED VGKVLAM MKERGVSIGV STHNIRIQSL CKRKRSGEAK ALLDGMLSSG MKPNAVTYGH LIHGFCNEGE FDEAKKLFKA MVN RGCKPD SECYFTLVYY LCKGGDFEAA LSLCKESMEK NWVPSFSIMK SLVNGLAKDS KVEEAKELIA QVKEKFTRNV ELWN EVEAA LPQ |
+Macromolecule #87: mS86
| Macromolecule | Name: mS86 / type: protein_or_peptide / ID: 87 Details: GWHPBJSH080930 Full sequence: MHYMGLFSRAGNIFRQPRALQASNAMLQGNLSLTPSKIFVGGLSPSTDVELLKEAFGSFG KIVDAVVVLDRESGLSRGFGFVTYDSIEVANNAMQAMQNKELDGRIIGVHPADSGGGGGG ...Details: GWHPBJSH080930 Full sequence: MHYMGLFSRAGNIFRQPRALQASNAMLQGNLSLTPSKIFVGGLSPSTDVELLKEAFGSFG KIVDAVVVLDRESGLSRGFGFVTYDSIEVANNAMQAMQNKELDGRIIGVHPADSGGGGGG GGFARRGGYGGGRGGYARGGFGRGGFGGGGYGFVR,Full sequence: MHYMGLFSRAGNIFRQPRALQASNAMLQGNLSLTPSKIFVGGLSPSTDVELLKEAFGSFG KIVDAVVVLDRESGLSRGFGFVTYDSIEVANNAMQAMQNKELDGRIIGVHPADSGGGGGG GGFARRGGYGGGRGGYARGGFGRGGFGGGGYGFVR Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.967962 KDa |
| Sequence | String: MHYMGLFSKA GNIFRQPRAM QASNAMLQGN LSLTPSKIFV GGLSPSTDVE LLKEAFGSFG KIVDAVVVLD RESGLSRGFG FVTYDSIEV ANNAMQAMQN KELDGRIIGV HPADSGGGGG GGGFARRGGY GGGRGGYARG GFGRGGFGGG GYGFVR |
+Macromolecule #88: uS2m
| Macromolecule | Name: uS2m / type: protein_or_peptide / ID: 88 / Details: GWHPBJSH046906 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.743545 KDa |
| Sequence | String: MTLHGAVIQK LLNTGSHLGR RAAEHHFKQY AYGTRNGMTI IDSDKTLICL RSAASFVANL ASARGNIFFV NTNPLFDEIV ELTSRRIQG DAYNHNRSTN LWKMGGFLTN SYSPKKFRSR HKKLCFGPTT MPDCVVVFDA ERKSSVVLEA AKLQIPVVAI V DPNVPLEF ...String: MTLHGAVIQK LLNTGSHLGR RAAEHHFKQY AYGTRNGMTI IDSDKTLICL RSAASFVANL ASARGNIFFV NTNPLFDEIV ELTSRRIQG DAYNHNRSTN LWKMGGFLTN SYSPKKFRSR HKKLCFGPTT MPDCVVVFDA ERKSSVVLEA AKLQIPVVAI V DPNVPLEF FEKITYPVPA RDSVKFVYLF CNVITKCFVA EQMKMGIKDG DLAA |
+Macromolecule #26: 26S rRNA
| Macromolecule | Name: 26S rRNA / type: rna / ID: 26 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 944.322562 KDa |
| Sequence | String: GAAUGCAUUG GAUGGAUGCC CGGGCAUUGA GAAGGAAGGA CGCUUUCAGA GGCGAAAGGC CAUGGGGAGA UACCGUCUGU GAUCCAUGG AUCUCCGAUC GGGAAACCGU AUCCAAGCUC CGUGGCUAGU CUGCGCUCUU UGGACUUUGA AAACUUAGCG A ACUGAAAC ...String: GAAUGCAUUG GAUGGAUGCC CGGGCAUUGA GAAGGAAGGA CGCUUUCAGA GGCGAAAGGC CAUGGGGAGA UACCGUCUGU GAUCCAUGG AUCUCCGAUC GGGAAACCGU AUCCAAGCUC CGUGGCUAGU CUGCGCUCUU UGGACUUUGA AAACUUAGCG A ACUGAAAC AUCUAAGUAG CUAAAGGAAG GGAAAUCAAC CGAGACCCCG UUAGUAGCGG CGAGCGAGAG CGGAUUUGGG AU UUGAAGA AAAAAAAAAA AAAUUUUUUU UAAAAAAUUU UUUUUUUUAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAU UUUUUU UUUUUUUUUA CUGUAAUUGU GAAAAGGUUG GAAGAUCUGG CCAAAGAAGG UGAUAGCCCU GUAGAUUCGU UCCU AUUUU UCGAUCCUUC CCAGUAAAAC GCGGCGUGUU CGAAUUCUGA UCGCUUUUAC GCGAGAAAGG GGGACCACCC UCUAA GCCU AAGUAUUCCU CAAUGACCGA UAGCGUACAA GUACCGUGAG GGAAAGGUGA AAAGAACCCU AUGACGGGAG UGCAAU AGA GAACCUGAGA UCCGAUGCGA ACAAUCAGUC GAAGGAGUAG UCAAGCGCAC UCACUCUAAC GGCGUACCUU UUGCAUG AU GGGUCAGCGA GGAAAUGGGA AGAGCGGCUU AAGCCAUUAG GUGUAGGCGC UUUCCAAAGG UGGAAUCUUC UAGUUCUU C CUAUUUGACC CGAAACCGAU CGAUCUAGCC AUGAGCAGGU UGAAGAGAGC UCUACAGGCC UUGGAGGACC GAACCCACG UAUG(PSU)GGCAA AAUACGGGGA UGACUUGUGG CUAGGGGUGA AAGGCCAACC AAGAUCGGAU AUAGCUGGUU UUCCGC GAA AUCUAUUUCA GUAGAGCGUA UGAUGUCGAU GGCCCGAGGU AGAGCACUCA AUGGGCUAGG GUGCUUACCA ACCCCAG GG AAACUCCGAA UACAGGCCGU UCUCGUUUGU ACAGACAGAC UUUUGGGG(PSU)G CUAAGAUCCA AAGUCGAGAG GGAA ACAGC CCAGAUCGUA CGCUAAGGUC CCUAAGCAAU CACUUAGUGG AAAAGGAAGU GAUCGAGCGA UGACAACCAG GAGGU GGGC UUGGAAGCAG CCAUCCUUUG AAGAAAGCGU AAUAGCUCAC UGGUCUAGCU CCAUGGCACC GAAAAUGUAU CAGGGC UCA AGUGAUUCAC CGAAGCGACG AGACCUUGCA AGCUGCUUUU CCAAGUGUCA GUAGCGGAAC GUUCUGUCAA UCGGAGA AG GUUUUUGGUG ACAAGACCUG GAGAUAUCAG AAGUGAGAAU GCUGACAUGA GUAACGAAAA AUCCUGUGAA AAACAAGA U CGCCUGCCAG UGGAAGGCUU UCUGCGUUCA GUCAAUCUAC GCAGAGUGAA UCGGUCCCUA AGGAAACCCC GAAAGGGCU GCCGUCCGAU GGGUACACGA AAGUGACGAA GUUGCUUUGA CUACAAAACC AUGCCUCUCU CUUGGAGCGA AUUGGAUGAU CGGGCCGAG GGCAGCGUAG CGCCUCUUCC CCUCACUCUC CUUUCCCUAA UAUGACUUGA GUCAUCAAAG CCUUUCUGAC U CAAAAAUU UUUUAAAAAA AAAAGUCGCG UAUUUUAAUA UCGCGACUCU AAUUUUUAGU CAAGAAGGUU GAAACUUCCA GG AAAAAAC UUCGAAUUGG GAGGGCGAUC CUCCCGGUGA ACUGACCGUA CCCCAAACCG ACACAGGUGA ACAAGUAGAG UAU ACUAGG GCGCUUGAGA GAACCAUGUC GAAGGAACUC GGCAAAAUGA CCCCGUAACU UCGGGAGAAG GGGUGCUCUC CUAU CUUUU GAUUAGGAAA GCGGCACAUA CCAGGGGGUA GCGACUGUUU AUUAAAAACA CAGGACUCUG CUAAGUGGUA ACACG AUGU AUAGAGUCUG ACACCUGCCC GGUGCUGGAA AGUCAAAAGG AGAAGUGUUA UAAGCUUUGA AUGGAAGCCC CGGUAA ACG GCGGCAGUAA CUCUAACUGU CCUAAGGUAG CGAAA(5MU)UCCU UGUCGCAUAA GUAGCGACCU GCACGAAUGG UGU AACGAC UGCCCCGCUG UCUCCGACAU GGACCCAGUG AAAUUGAAUU CUCCGUGAAG AUGCGGAGUA CCAACGGCUA GACG GUAAG ACCCCGUGCA CCUUCACUAU AGCUUCGCAG UGACAACCUU GAUCGAAUGU GUAGGAUAGG UGGGAGGUCC UCACA UAGA AAGACCAAUC CUGAAAGACC ACUCUUUCGU CUAAGGGUGC CUAACCGCCG CGGCGGGACA CUGCGAGGUG GGUAGU UUA UCUG(OMG)GGCGG AUGCCUCCUA AAGAGUAACG GAGGUGUGCG AAGGUAGGCU CAAGCAAAUC UGCUCGUGAG CGU AAUGGU AUAAGCCUGC CUGACUGUGA GACCGACUGG UCGAACAGAG ACGAAAGUCG GCCAUAGUGA UCCGGGAGUC CCGU GUGGA AGGGCUCUCG CUCAACGGAU CAAAGGUACG CCGGGGA(H2U)AA CAGGC(PSU)GAUG ACUCCCAAGA GCUCUUA UC GACGGAGUCG UUUGGCA(OMC)CU CG(2MA)UGUCGAC UCAUCACAUC CUGGGGUUGA AGAAGGUCCC AAGGGUUCGG U(OMU)GUUCGCC GAUUCAAGUG GUACGUGAGU (PSU)GGGUUUAGA ACGUCGUGAG ACAGU(PSU)CGGU UCCUAUCUA CCGUUGGUGU UAAAGCGAGA ACUGCGAGGA GCCAACCCUA GUACGAGAGG ACUGGGUUGG GCCAACCUAU GGUGUACCGG UUGUUAUGA CAAUAGCAGC GCCGGGAAGC UAAGUUGGUA UGGAAGAACU GCUGCUUAGC GGGAAAUCCU UCUCUAUACA A GUUCUCGG AACAGGUUUU AGAACAGAAC UUCGAUAGGC GAGAGGUGGA AGCACCGCGA GGUGUGGAGC CAUCUCGUAC UA AACGAA |
+Macromolecule #27: 5S rRNA
| Macromolecule | Name: 5S rRNA / type: rna / ID: 27 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.992594 KDa |
| Sequence | String: AAACCGGGCA CUACGGUGAG ACGUGAAAAC ACCCGAUCCC AUUCCGACCU CGAUAUGUGG AAUCGUCUUG CGCCAUAUGU ACUGAGAUU GUUCGGGAGA CAUGGUCCAA GCCCGGUGA |
+Macromolecule #75: tRNA
| Macromolecule | Name: tRNA / type: rna / ID: 75 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.541719 KDa |
| Sequence | String: GCGGGG(4SU)GGA GCAGCCUGGU AGCUCGUCGG G(5MC)UCAUAACC CGAAGGUCGU CGG(5MU)(PSU)CAAAU CCGG CCCCC GCAACCA |
+Macromolecule #76: 18S rRNA
| Macromolecule | Name: 18S rRNA / type: rna / ID: 76 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 514.869688 KDa |
| Sequence | String: CAUAGUCAAA AGAAGAGUUU GAUUCUGGCU CAGAAGGAAC GCUAGCUAUA UGCUUAACAC AUGCAAGUCG AACGUUGUUC UAAAAAAAA AAAAAAUUUU UUUUUUUUUA AAAAAAAAAA AAAUUUUUUU UAAGAGUUGA GAACAAAGUG GCGAACGGGU G AGUAAGGC ...String: CAUAGUCAAA AGAAGAGUUU GAUUCUGGCU CAGAAGGAAC GCUAGCUAUA UGCUUAACAC AUGCAAGUCG AACGUUGUUC UAAAAAAAA AAAAAAUUUU UUUUUUUUUA AAAAAAAAAA AAAUUUUUUU UAAGAGUUGA GAACAAAGUG GCGAACGGGU G AGUAAGGC GUGGGAAUCU GCCGAACAGU UCGGGCUAAA UCCCGAAGAA AGCUAGAAAG CGCUGUUUGA UGAGCCUGCG UA GUAUUAG GUAGUUGGUU AGGUAAAGGC UGACCAAGCC GAUGAUGCUU AGCUGGUCUU UUCGGACGAU CAGCCACACC GGG ACUGAG ACACGGCCCG GACUCCCACG GGGGGCAGCA GUGGGGAAUC UUGGACAAUG GGCGCAAGCC UGAUCCAGCA AUAU UGCGU GAGUGAAGAA GGGCAAUUCC GCUCGUAAAG CUCUUUCGUC GAGUGCGCGA UCAUGACAUG ACUCGAGGAA GAAGC CCCG GCUAACUCCG UGCCAGCAGC C(G7M)CGGUAAAA CGGGGGGGGC AAGUGUUCUU CGGAAUGACU AGGCGUAAAG GG CACGUAG GCGGUUCAUC GGGUUGAAAG UGAAAGUCGU CAAAAAGUGG CGGAAUGCUU UCGAAACCAC UGAACUUGAG UGA GACAGA GGAGAGUGGA AUUUCGUGUG GAGGGGUGAA AUCUACGGAU CUACGAAGGA ACGCCAAAAG CGAAGGCAGC UCUC UGGGU CUCUACCGAC GCUGGUGUGC GAAAGCAUGG GGAGCGAACG GGAUUAGAUA CCCUGGUAGU CCAUGCCGUA AACGA UGAG UGUUCGCCCU UGGUCUAUUC UGAUCUGGGG CUCAGCUAAC GCGUUAAACA CUCCGCCUGG GGAGUACGGU CGCAAG ACC AAAACUCAAA GGAAUUGACG GGGGCCUGCA CAAGCGGUGG AGCAUGUGGU UUAAUUCGAU GCAACGCGCA GAACCUU AC CAGCCCUUGA CAUAUGAACG AGAAACCCUA UCCUUAACGG GAUGGUACUG ACUUUCAUAC AGGUGCUGCA UGGCUGUC G UCAGCUCGUG UCGUGAGAUG UUUGGUUAAG UCCUAUAACG AGCGAAACCC UCGCCUUGUG UUGCUCAGAC AUGCGCCUA AGGAGAACUC CGAAACAAAC AAAAAGGUGC GUGACGCACU CACGAGGGAC UGCCAGUGAU AUACUGGAGG AAGGUGGGGA UGACGUCAA GUCCGCAUGG CCCUUAUGGG CUGGGCCACA CACGUGCUAC AAUGGCAAUU ACAAUGGGAA GCAAGGCUGU A AGGCGGAG CGAAUCCGGA AAGAUUGCCU CAGUUCGGAU UGUUCUCUGC AACUCGGGAA CAUGAAGUUG GAAUCGCUAG UA AUCGCGG AUCAGCAUGC CGCGGUGAAU AUGUACCCGG GCCCUGUACA CACCG(4OC)CCGU CACACCCUGG GAAUUGGUU UCGCCCGAAG CAUCGGACCA AUGAUCACCC AGCACUUCUG UGUAACACUA GUCACACAAA GACUUUUGGU GGUCUUAUUG ACGCAUACC ACGGUGGGGU CUUCGACUGG GGUGAAGUCG (UR3)AACAAGGUA GCCGUAGG(2MG)G (MA6)(MA6)CCU GUGG CUGGAUUAUC C |
+Macromolecule #77: mRNA
| Macromolecule | Name: mRNA / type: rna / ID: 77 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.907237 KDa |
| Sequence | String: AAAAUA |
+Macromolecule #89: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 89 / Number of copies: 357 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
+Macromolecule #90: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 90 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
+Macromolecule #91: CHLORAMPHENICOL
| Macromolecule | Name: CHLORAMPHENICOL / type: ligand / ID: 91 / Number of copies: 1 / Formula: CLM |
|---|---|
| Molecular weight | Theoretical: 323.129 Da |
| Chemical component information | 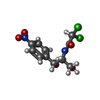 ChemComp-CLM: |
+Macromolecule #92: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 92 / Number of copies: 74 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
+Macromolecule #93: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 93 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-9evs: |
+ About Yorodumi
About Yorodumi
-News
-Feb 9, 2022. New format data for meta-information of EMDB entries
New format data for meta-information of EMDB entries
- Version 3 of the EMDB header file is now the official format.
- The previous official version 1.9 will be removed from the archive.
Related info.: EMDB header
EMDB header
External links: wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
EMDB accession codes are about to change! (news from PDBe EMDB page)
- The allocation of 4 digits for EMDB accession codes will soon come to an end. Whilst these codes will remain in use, new EMDB accession codes will include an additional digit and will expand incrementally as the available range of codes is exhausted. The current 4-digit format prefixed with “EMD-” (i.e. EMD-XXXX) will advance to a 5-digit format (i.e. EMD-XXXXX), and so on. It is currently estimated that the 4-digit codes will be depleted around Spring 2019, at which point the 5-digit format will come into force.
- The EM Navigator/Yorodumi systems omit the EMD- prefix.
Related info.: Q: What is EMD? /
Q: What is EMD? /  ID/Accession-code notation in Yorodumi/EM Navigator
ID/Accession-code notation in Yorodumi/EM Navigator
External links: EMDB Accession Codes are Changing Soon! /
EMDB Accession Codes are Changing Soon! /  Contact to PDBj
Contact to PDBj
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
-Yorodumi
Thousand views of thousand structures
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































